explore_mtsplice
2022-09-20
Last updated: 2022-10-07
Checks: 6 1
Knit directory: funcFinemapping/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210404) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 9e4e697. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .ipynb_checkpoints/
Ignored: analysis/.Rhistory
Ignored: analysis/PTR_m6A.nb.html
Ignored: analysis/build_annotations_for_single_cell_data.nb.html
Ignored: analysis/lab4_prepare.nb.html
Ignored: analysis/ldsc_results.nb.html
Ignored: analysis/learn_archR.nb.html
Ignored: analysis/mtsplice_finemapping_results.nb.html
Ignored: analysis/results.nb.html
Ignored: analysis/snp_finemapping_results.nb.html
Ignored: analysis/splicing.nb.html
Ignored: analysis/susie_tutorial.nb.html
Untracked files:
Untracked: SCZ_pval_vs_MAF.png
Untracked: SNPs_categories,png
Untracked: SNPs_categories.png
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/PTR_m6A.Rmd
Untracked: analysis/PTR_splicing_mtsplice.Rmd
Untracked: analysis/PTR_splicing_spliceAI.Rmd
Untracked: analysis/explore_mtsplice.Rmd
Untracked: analysis/snp_finemapping_spliceAI_prior.Rmd
Untracked: analysis/snp_finemapping_with_priors.Rmd
Untracked: bmi_locus1410.pdf
Untracked: code/.ipynb_checkpoints/
Untracked: code/.snakemake/
Untracked: code/Euro_LD_Chunks.RData
Untracked: code/Snakefile
Untracked: code/config.yaml
Untracked: code/environment.yml
Untracked: code/extract_mmsplice_predictions_bychr.R
Untracked: code/extract_mtsplice_maxPred.sh
Untracked: code/get_indiv_annotation_from_predictions.R
Untracked: code/heatmap_torus_enrichment.ipynb
Untracked: code/ldsc.log
Untracked: code/ldsc.results
Untracked: code/ldsc_regression.sh
Untracked: code/make_joint_annotations.R
Untracked: code/make_plots.R
Untracked: code/out/
Untracked: code/plot_torus_enrichment.R
Untracked: code/prepare_torus_inputs.R
Untracked: code/run_ldsc.sh
Untracked: code/run_ldsc_with_bed.sh
Untracked: code/run_ldsc_with_bed_v2.sh
Untracked: code/run_susie.R
Untracked: code/run_susie_in_parallel.R
Untracked: code/run_torus.sh
Untracked: code/run_torus/
Untracked: code/sctype/
Untracked: code/slurm-22619162.out
Untracked: code/slurm-22619163.out
Untracked: code/slurm-22619164.out
Untracked: code/slurm-22619165.out
Untracked: code/slurm-22619166.out
Untracked: code/slurm-22619199.out
Untracked: code/slurm-22619200.out
Untracked: code/slurm-22619201.out
Untracked: code/slurm-22619202.out
Untracked: code/slurm-22619203.out
Untracked: code/slurm-22619228.out
Untracked: code/slurm-22619229.out
Untracked: code/slurm-22619230.out
Untracked: code/slurm-22619231.out
Untracked: code/slurm-22619232.out
Untracked: code/slurm-22619234.out
Untracked: code/slurm-22619235.out
Untracked: code/slurm-22619236.out
Untracked: code/slurm-22619237.out
Untracked: code/slurm-22619238.out
Untracked: code/slurm-22619289.out
Untracked: code/slurm-22619290.out
Untracked: code/split_vcf.sh
Untracked: code/test_mtsplice.txt.gz
Untracked: code_backup/
Untracked: data/ASD_denovo_variants.1.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.10.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.11.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.12.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.13.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.14.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.15.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.16.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.17.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.18.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.19.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.2.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.20.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.21.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.22.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.3.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.4.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.5.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.6.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.7.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.8.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.9.sorted.vcf.gz
Untracked: data/ASD_denovo_variants.vcf.gz
Untracked: data/ScTypeDB_full.xlsx
Untracked: data/compare_spliceai_and_mmsplice.RData
Untracked: data/header.txt.gz
Untracked: data/hg19_gtf_genomic_annots_ver2.gr.rds
Untracked: data/mmsplice_mtsplice_cutoffs.M5.txt
Untracked: data/mmsplice_mtsplice_cutoffs.txt
Untracked: data/num_overlaps_finemapped_SNPs_and_ctcf.txt
Untracked: data/process_vcf.sh
Untracked: data/qqplot_SNPs_high_spliceAI.png
Untracked: data/qqplot_SNPs_high_spliceAI_scores_SCZ.png
Untracked: data/qqplot_SNPs_high_spliceAI_scores_aFib.png
Untracked: data/qqplot_SNPs_high_spliceAI_scores_allergy.png
Untracked: data/spliceAIandMAF.txt.gz
Untracked: data/torus_enrichment_novel_annot.est
Untracked: data/torus_joint_enrichment.est
Untracked: data/torus_joint_refined_enrichment.est
Untracked: data/vcf/
Untracked: enhancer_gene_feature.rmd
Untracked: fig1_panels.pdf
Untracked: fig2.pdf
Untracked: fig_panel2.pdf
Untracked: gene_mapping.pdf
Untracked: output/AAD/GMP_merge_stats.txt
Untracked: output/AAD/Wang2020_joint.results
Untracked: output/AAD/Wang2020_joint_T.results
Untracked: output/AAD/Wang2020_joint_tissueResT.results
Untracked: output/AAD/allergy/Ulirsch2019/GMP_merge_compare_old.est
Untracked: output/AAD/allergy/Ulirsch2019_disjoint_snps.sumstats
Untracked: output/AAD/allergy/Wang2020_T_subsets.est
Untracked: output/AAD/allergy/Wang2020_T_subsets_indiv.est
Untracked: output/AAD/allergy/Wang2020_T_tissueRes.est
Untracked: output/AAD/allergy/Wang2020_joint_T.results
Untracked: output/AAD/allergy/Wang2020_joint_tissueResT.results
Untracked: output/AAD/allergy/Wang2020_tissueResT.est
Untracked: output/AAD/allergy/torus_enrichment_CD4.est
Untracked: output/AAD/allergy/torus_enrichment_CD8.est
Untracked: output/AAD/allergy/torus_enrichment_non_tissueRes_T.est
Untracked: output/AAD/allergy/torus_enrichment_tissueMigraT.est
Untracked: output/AAD/allergy/torus_enrichment_tissueResT_C6.est
Untracked: output/AAD/allergy/torus_enrichment_tissueResT_C8.est
Untracked: output/AAD/allergy/torus_enrichment_tissueRes_T.est
Untracked: output/AAD/allergy/torus_enrichment_tissueResident_T_cells.est
Untracked: output/AAD/asthma_adult/Ulirsch2019/CD4_compare_old.est
Untracked: output/AAD/asthma_adult/Ulirsch2019/CD8_compare_old.est
Untracked: output/AAD/asthma_adult/Ulirsch2019/GMP_merge_compare_old.est
Untracked: output/AAD/asthma_adult/Wang2020_T_subsets.est
Untracked: output/AAD/asthma_adult/Wang2020_T_subsets_indiv.est
Untracked: output/AAD/asthma_adult/Wang2020_T_tissueRes.est
Untracked: output/AAD/asthma_adult/Wang2020_joint_T.results
Untracked: output/AAD/asthma_adult/Wang2020_joint_tissueResT.results
Untracked: output/AAD/asthma_adult/torus_enrichment_CD4.est
Untracked: output/AAD/asthma_adult/torus_enrichment_CD8.est
Untracked: output/AAD/asthma_adult/torus_enrichment_non_tissueRes_T.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueMigraT.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueResT_C6.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueResT_C8.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueRes_T.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueResident_T_cells.est
Untracked: output/AAD/asthma_child/CD4_compare.est
Untracked: output/AAD/asthma_child/CD8_compare.est
Untracked: output/AAD/asthma_child/Ulirsch2019/GMP_merge_compare_old.est
Untracked: output/AAD/asthma_child/Ulirsch2019/torus_enrichment_CD4.est
Untracked: output/AAD/asthma_child/Ulirsch2019/torus_enrichment_CD8.est
Untracked: output/AAD/asthma_child/Wang2020_T_subsets.est
Untracked: output/AAD/asthma_child/Wang2020_T_subsets_indiv.est
Untracked: output/AAD/asthma_child/Wang2020_T_tissueRes.est
Untracked: output/AAD/asthma_child/Wang2020_joint_T.results
Untracked: output/AAD/asthma_child/Wang2020_joint_tissueResT.results
Untracked: output/AAD/asthma_child/torus_enrichment_CD4.est
Untracked: output/AAD/asthma_child/torus_enrichment_CD8.est
Untracked: output/AAD/asthma_child/torus_enrichment_non_tissueRes_T.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueMigraT.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueResT_C6.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueResT_C8.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueRes_T.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueResident_T_cells.est
Untracked: output/LDL_ukb_L10.gif
Untracked: output/LDL_ukb_L10.pdf
Untracked: output/background_SNPs_annotated_percent.txt
Untracked: output/ldsc
Untracked: output/locus_1452.gif
Untracked: output/locus_1452.pdf
Untracked: output/spliceAI_vs_MAF.png
Untracked: output/splicing/.ipynb_checkpoints/
Untracked: output/splicing/PTR_across_traits_annotations.results
Untracked: output/splicing/PTR_joint.pdf
Untracked: output/splicing/PTR_joint.png
Untracked: output/splicing/QQplot_mmsplice.png
Untracked: output/splicing/QQplot_mmsplice_top15.png
Untracked: output/splicing/QQplot_mmsplice_vs_mtsplice.png
Untracked: output/splicing/QQplot_mmsplice_vs_mtsplice_SCZ.png
Untracked: output/splicing/TSplice_scores_distribution.png
Untracked: output/splicing/aFib_joint_comparison.pdf
Untracked: output/splicing/allergy_joint_comparison.pdf
Untracked: output/splicing/header.txt
Untracked: output/splicing/m6A_enrichment_across_traits.pdf
Untracked: output/splicing/mmsplice_vs_mtsplice_AA_heart.png
Untracked: output/splicing/mmsplice_vs_mtsplice_Hypothalamus_brain.png
Untracked: output/splicing/mmsplice_vs_mtsplice_LV_heart.png
Untracked: output/splicing/mmsplice_vs_mtsplice_across_tissues.png
Untracked: output/splicing/mtsplice_enrichment_across_traits.pdf
Untracked: output/splicing/prior/
Untracked: output/splicing/scz_PTR_annotations.results
Untracked: output/splicing/scz_joint_comparison.pdf
Untracked: output/splicing/scz_neuOCR_m6a_DMR.results
Untracked: output/splicing/scz_spliceAI0.03_hist.png
Untracked: output/splicing/scz_spliceAI0.03_scatterplot.png
Untracked: output/splicing/scz_spliceai_binary0.03.results
Untracked: output/splicing/summarizing_aFib_PIPs.pdf
Untracked: output/splicing/torus_afib_spliceai.est
Untracked: output/splicing/torus_annotations_spliceai0.01.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.03.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.05.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.07.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.1.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.2.txt.gz
Untracked: output/splicing/torus_annotations_spliceai9.txt.gz
Untracked: output/splicing/torus_enrichment_joint_scz_mtsplice0.6_hypothalamus-brain.est
Untracked: output/splicing/torus_enrichment_joint_scz_spliceAI.est
Untracked: output/splicing/torus_spliceai0.01.enrichment
Untracked: output/splicing/torus_spliceai0.03.enrichment
Untracked: output/splicing/torus_spliceai0.05.enrichment
Untracked: output/splicing/torus_spliceai0.07.enrichment
Untracked: output/splicing/torus_spliceai0.1.enrichment
Untracked: output/splicing/torus_spliceai0.2.enrichment
Untracked: output/splicing/torus_zscores.txt.gz
Untracked: output/torus
Untracked: panel_figure2.pdf
Untracked: test.txt
Unstaged changes:
Deleted: .Rprofile
Modified: analysis/lab4_prepare.Rmd
Modified: analysis/ldsc_PTR_results.Rmd
Deleted: output/AAD/Caldero2019_disjoint_snps.sumstats
Modified: output/AAD/allergy/Caldero2019_disjoint_snps.sumstats
Modified: output/AAD/allergy/Ulirsch2019/GMP_merge_compare.est
Modified: output/AAD/allergy/Wang2020_indiv.est
Modified: output/AAD/allergy/Wang2020_joint.results
Deleted: output/AAD/asthma/Caldero2019_diffDA_annot_percent.txt
Deleted: output/AAD/asthma/Caldero2019_stimuDA_annot_percent.txt
Deleted: output/AAD/asthma/celltype_specific_adult_lungs_torus.est
Deleted: output/AAD/asthma/diffe_adult_blood_torus.est
Deleted: output/AAD/asthma/joint_blood_immune_rest_vs_stimu.est
Deleted: output/AAD/asthma/joint_lung_vs_blood_immune_diff_torus.est
Deleted: output/AAD/asthma/joint_lung_vs_blood_immune_stimu_torus.est
Deleted: output/AAD/asthma/lung_clusters_dict.txt
Deleted: output/AAD/asthma/lung_clusters_info.txt
Deleted: output/AAD/asthma/stimu_adult_blood_torus.est
Deleted: output/AAD/asthma/torus_enrichment_all_rest.est
Deleted: output/AAD/asthma/torus_enrichment_all_stimulated.est
Deleted: output/AAD/asthma/zhang2021_annot_percent.txt
Deleted: output/AAD/asthma/zhang2021_cell_type_overlaps.txt
Deleted: output/AAD/asthma/zhang2021_peaks_per_celltype.txt
Modified: output/AAD/asthma_adult/Ulirsch2019/CD4_compare.est
Modified: output/AAD/asthma_adult/Ulirsch2019/CD8_compare.est
Deleted: output/AAD/asthma_adult/Ulirsch2019/GMP_merge_compare.est
Modified: output/AAD/asthma_adult/Wang2020_indiv.est
Modified: output/AAD/asthma_adult/Wang2020_joint.results
Modified: output/AAD/asthma_child/Ulirsch2019/GMP_merge_compare.est
Modified: output/AAD/asthma_child/Wang2020_indiv.est
Modified: output/AAD/asthma_child/Wang2020_joint.results
Deleted: output/asthma/Caldero2019_diffDA_annot_percent.txt
Deleted: output/asthma/Caldero2019_stimuDA_annot_percent.txt
Deleted: output/asthma/celltype_specific_adult_lungs_torus.est
Deleted: output/asthma/diffe_adult_blood_torus.est
Deleted: output/asthma/joint_lung_vs_blood_immune_diff_torus.est
Deleted: output/asthma/joint_lung_vs_blood_immune_stimu_torus.est
Deleted: output/asthma/lung_clusters_dict.txt
Deleted: output/asthma/lung_clusters_info.txt
Deleted: output/asthma/stimu_adult_blood_torus.est
Deleted: output/asthma/zhang2021_annot_percent.txt
Deleted: output/asthma/zhang2021_cell_type_overlaps.txt
Deleted: output/asthma/zhang2021_peaks_per_celltype.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/explore_mtsplice.Rmd) and
HTML (docs/explore_mtsplice.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 9e4e697 | Jing Gu | 2022-10-07 | understand mtsplice outputs |
| html | a2a8fae | Jing Gu | 2022-10-01 | explore MTSplice |
MMSplice/MTSplice predictions
MTSplice model \[ \Delta logit(\Psi_{e,t}) = logit(\Psi_{e,avg}^{ref} ) + \text{MMSplice}(S_{ref}-S_{alt}) + \text{TSplice}(S_{alt}, \text{tissue}) - logit(\Psi_{e,t}^{ref}) \] MMSplice model \[ \Delta logit(\Psi_{e,t}) = logit(\Psi_{e,avg}^{ref} ) + \text{MMSplice}(S_{ref}-S_{alt}) - logit(\Psi_{e,t}^{ref}) \]
Understand MTSplice outputs.
Three types of outputs
Tissue_ref: psi scores for ref allele in one tissue
Tissue_delta_psi: difference in psi scores between ref and alt
Tissue:logit psi difference between ref and alt
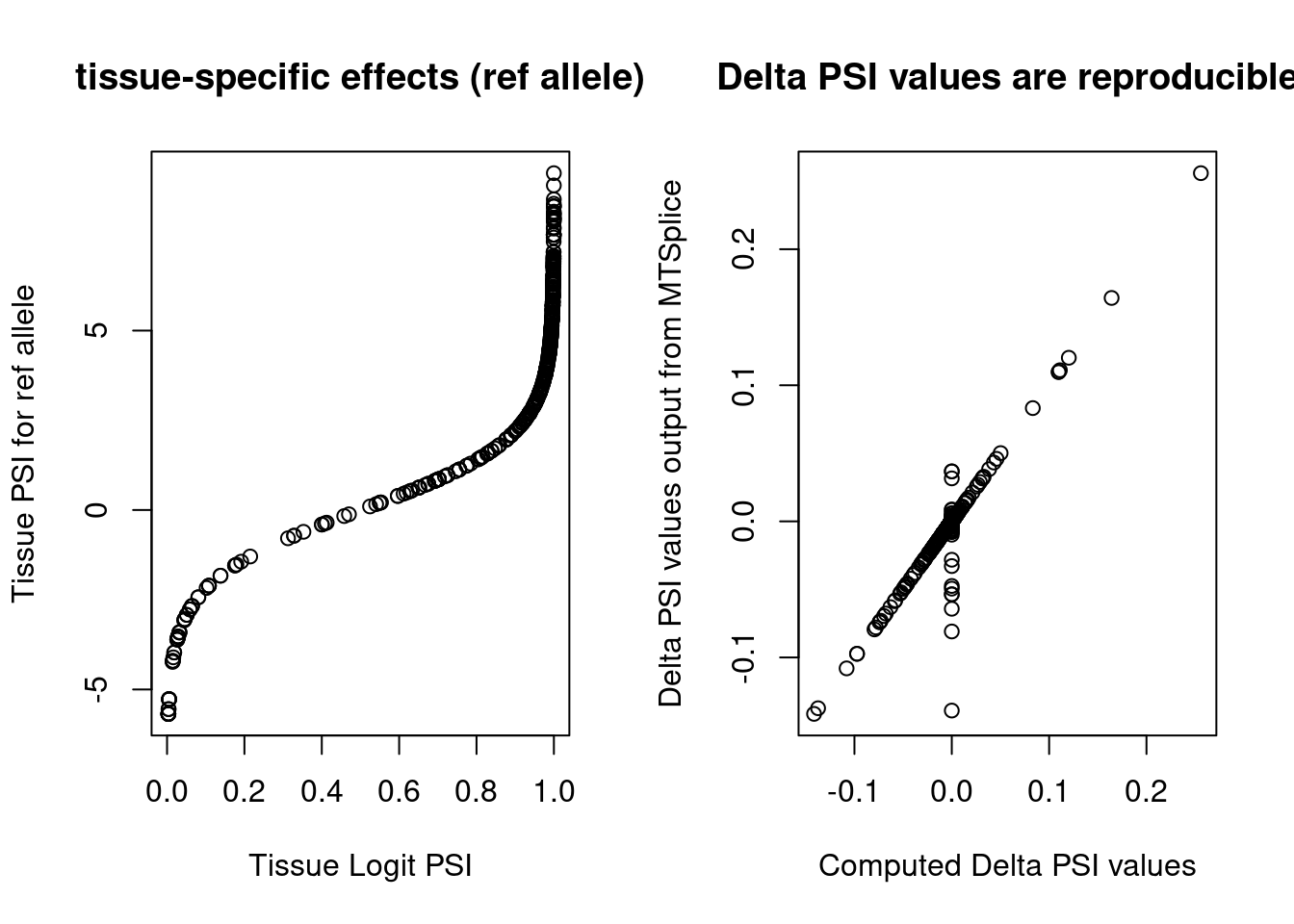
Version Author Date a2a8fae Jing Gu 2022-10-01
Distribution of the location of predicted SNPs
Warning in sprintf("%s%% predicted SNPs are within 5kb from the nearest exon", :
one argument not used by format '%s%% predicted SNPs are within 5kb from the
nearest exon'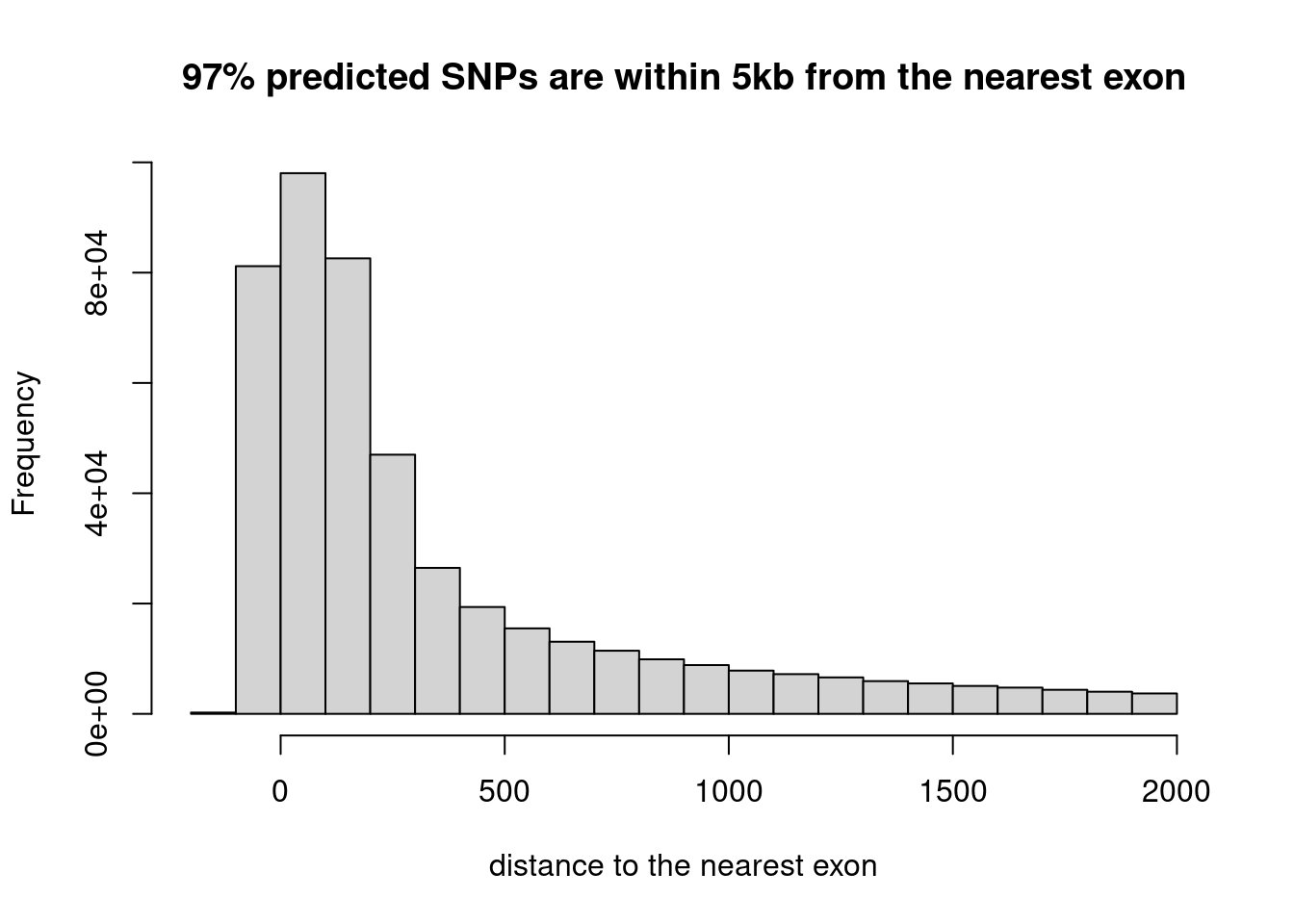
| Version | Author | Date |
|---|---|---|
| a2a8fae | Jing Gu | 2022-10-01 |
Majority of MTSplice predicted SNPs are within 500 bp from the nearest exon.
MMSplice predictions on ASD de novo mutations
Replicate the results in fig 7a using the reported de novo mutations from ASD patients (Zhou et al.) The de novo mutations were filtered by 1) within exons or their 300-nt flanking intronic regions and 2) having MMSplice predicted \(\Delta\text{logit}(\Psi)\) with a magnitude greater than 0.05.
Legend:
- The color specifies whether an individual is a proband or sibling.
- The dot on the plot represents mean predicted scores and error bar stands for 95% confidence interval
[1] "The number of mutations that satisfy the criteria is 3725, very close to 3884 mutations found in the paper."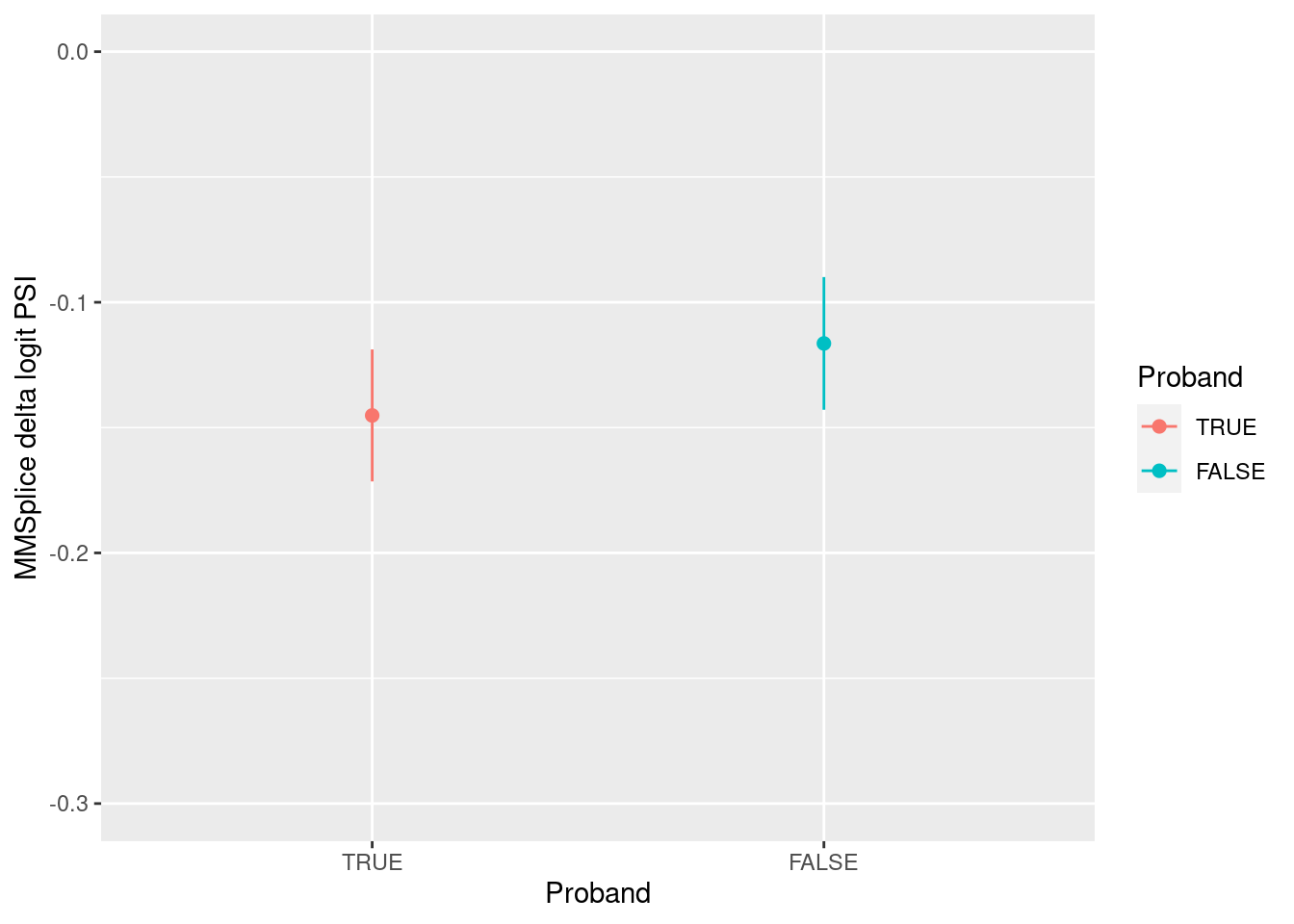
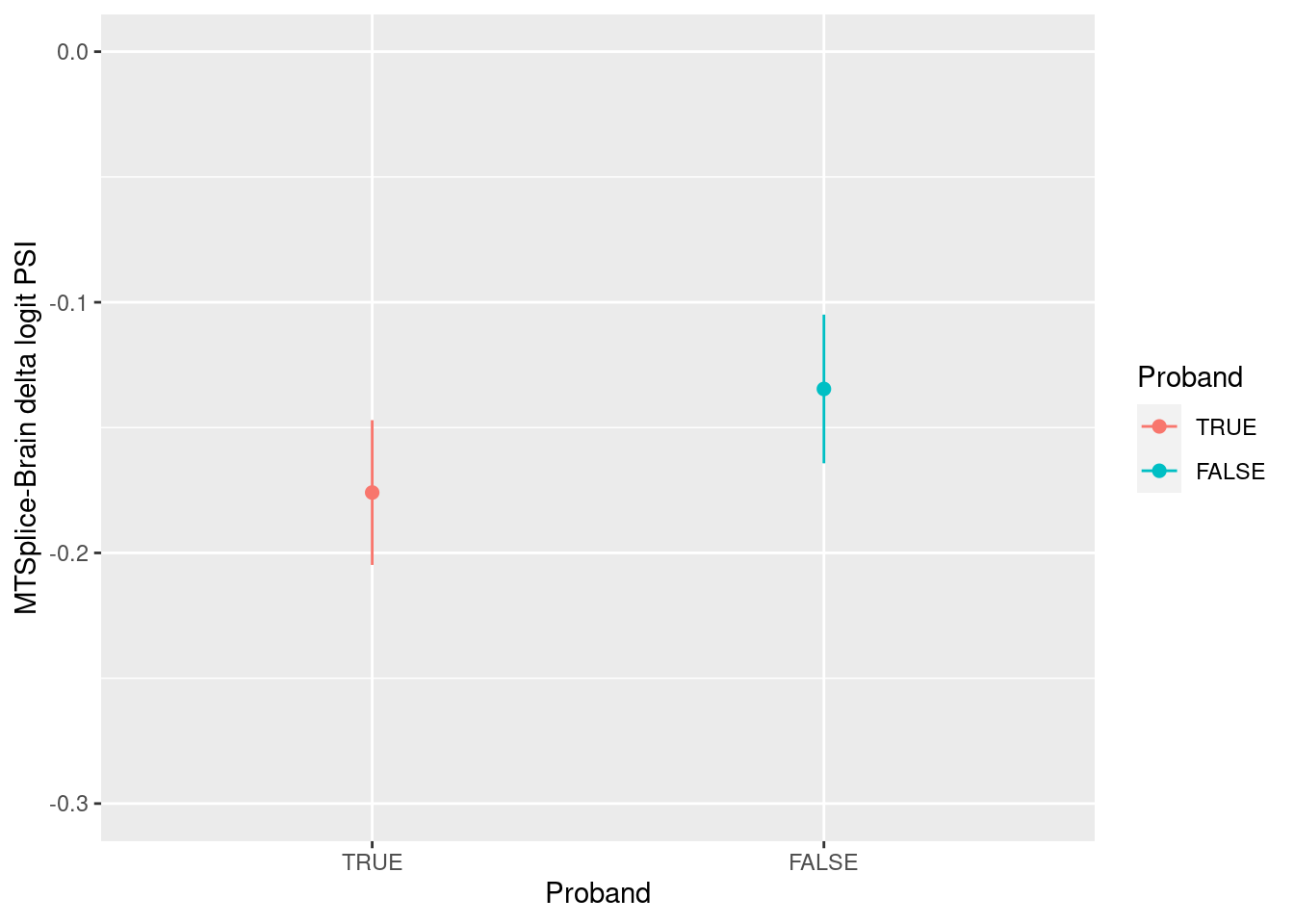
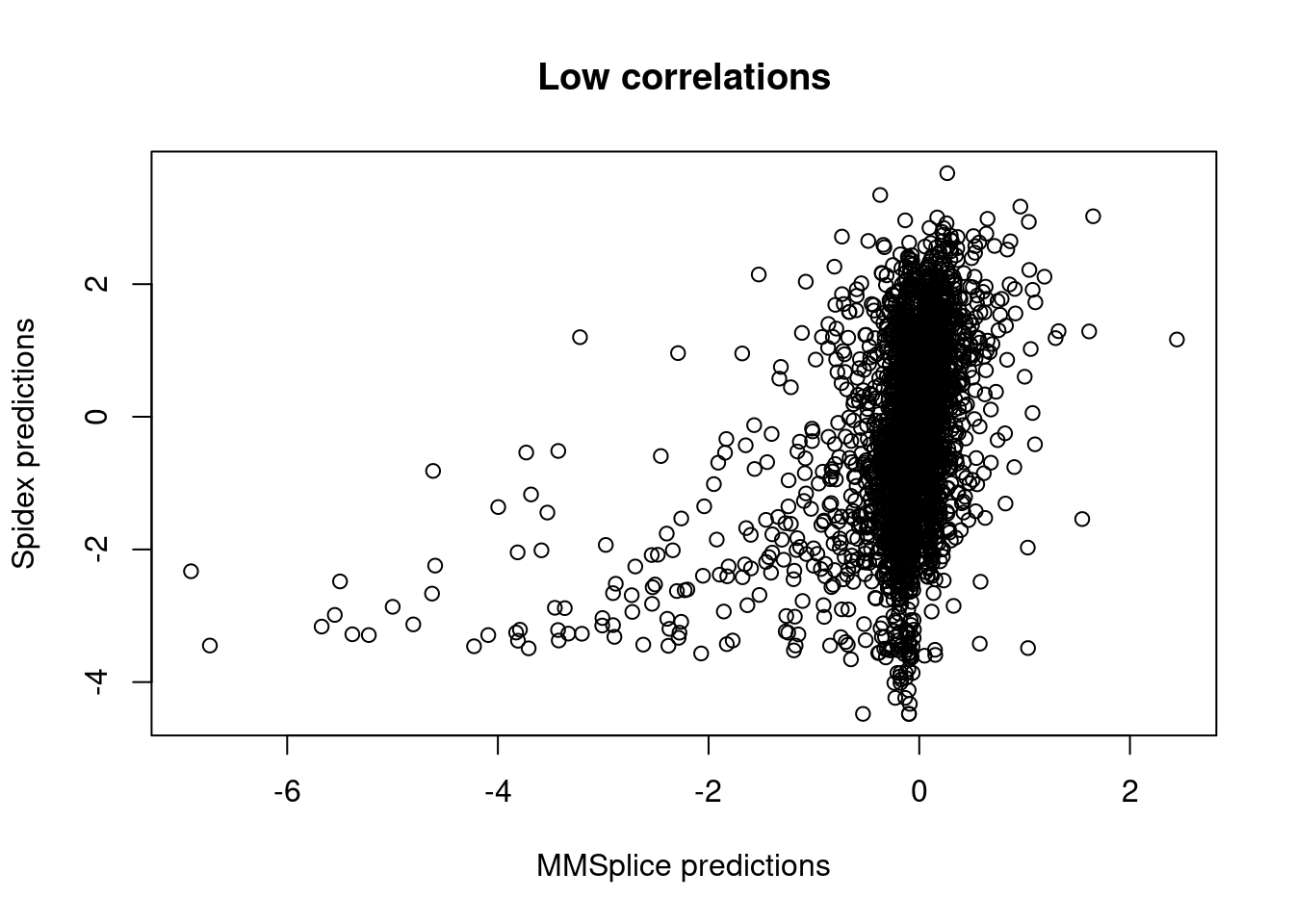
Compare with Spidex results
data directory: /home/jinggu/cluster/data/features/raw/splicing/spidex
Around 3000 out of 3700 de novo variants with splicing
altering effects were found in Spidex predictions.
| Version | Author | Date |
|---|---|---|
| a2a8fae | Jing Gu | 2022-10-01 |
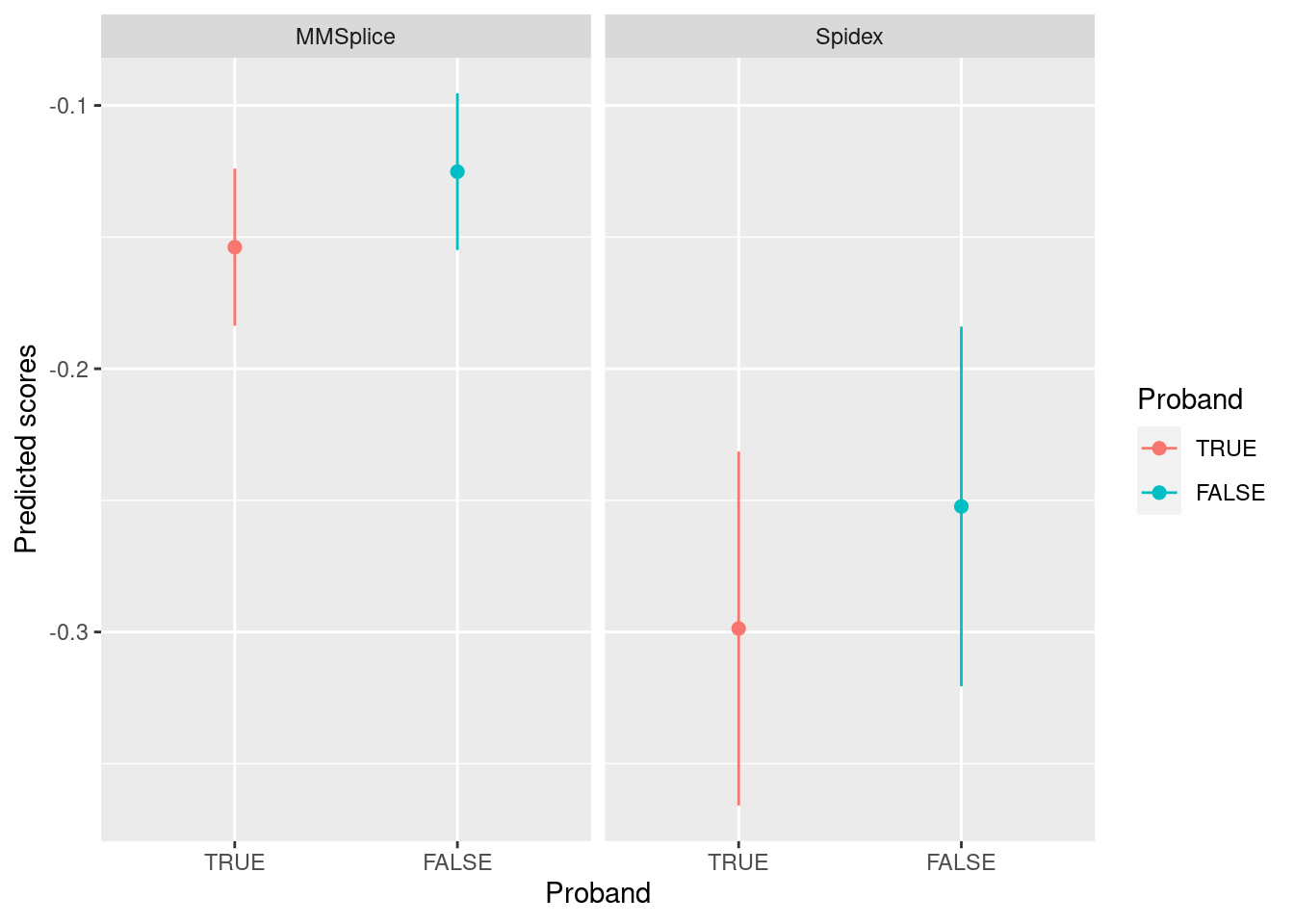
In our analysis, MMSplice or Spidex now shows comparable results as the ones displayed on Fig 7a from the paper. The main difference is confidence interval was drawn instead of standard deviation.
sessionInfo()R version 4.2.0 (2022-04-22)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=C LC_MONETARY=C LC_MESSAGES=C
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggplot2_3.3.6 dplyr_1.0.9 data.table_1.14.2
loaded via a namespace (and not attached):
[1] tidyselect_1.1.2 xfun_0.30 bslib_0.3.1 reshape2_1.4.4
[5] purrr_0.3.4 colorspace_2.0-3 vctrs_0.4.1 generics_0.1.2
[9] htmltools_0.5.2 yaml_2.3.5 utf8_1.2.2 rlang_1.0.2
[13] R.oo_1.24.0 jquerylib_0.1.4 later_1.3.0 pillar_1.7.0
[17] glue_1.6.2 withr_2.5.0 DBI_1.1.2 R.utils_2.11.0
[21] plyr_1.8.7 lifecycle_1.0.1 stringr_1.4.0 munsell_0.5.0
[25] gtable_0.3.0 workflowr_1.7.0 R.methodsS3_1.8.1 evaluate_0.15
[29] labeling_0.4.2 knitr_1.39 fastmap_1.1.0 httpuv_1.6.5
[33] fansi_1.0.3 highr_0.9 Rcpp_1.0.8.3 promises_1.2.0.1
[37] scales_1.2.0 jsonlite_1.8.0 farver_2.1.0 fs_1.5.2
[41] digest_0.6.29 stringi_1.7.6 rprojroot_2.0.3 grid_4.2.0
[45] cli_3.3.0 tools_4.2.0 magrittr_2.0.3 sass_0.4.1
[49] tibble_3.1.7 crayon_1.5.1 whisker_0.4 pkgconfig_2.0.3
[53] ellipsis_0.3.2 assertthat_0.2.1 rmarkdown_2.14 rstudioapi_0.13
[57] R6_2.5.1 git2r_0.30.1 compiler_4.2.0