cTWAS_analysis_for_BMI
Jing Gu
2023-08-25
Last updated: 2023-08-25
Checks: 6 1
Knit directory: m6A_in_disease_genetics/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230331) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/projects/m6A_in_disease_genetics/code/ctwas/ctwas_config_b37.R | code/ctwas/ctwas_config_b37.R |
| ~/projects/m6A_in_disease_genetics/code/ctwas/qiansheng/locus_plot.R | code/ctwas/qiansheng/locus_plot.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d0f5634. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: analysis/m6A_switch_to_disease_h2g.nb.html
Ignored: data/plots/
Untracked files:
Untracked: HMGCR_locus_gene_tracks.pdf
Untracked: Rplots.pdf
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/IBD_E_S_m6A.Rmd
Untracked: analysis/IBD_E_S_m6A_output.Rmd
Untracked: analysis/LDL_E_S_m6A.Rmd
Untracked: analysis/LDL_m6A_output.Rmd
Untracked: analysis/RA_m6A_output.Rmd
Untracked: analysis/WhiteBlood_WholeBlood_E_M.Rmd
Untracked: analysis/identify_m6A_mechanisms_with_finemapping.Rmd
Untracked: analysis/lymph_m6A_output.Rmd
Untracked: analysis/pre_weights_m6AQTL.txt
Untracked: analysis/rbc_E_S_m6A_output.Rmd
Untracked: analysis/rbc_m6A_output.Rmd
Untracked: analysis/summarize_ctwas_m6A_results.Rmd
Untracked: analysis/wbc_E_S_m6A_output.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/all_m6a_sites_with_paired_cisNATs_summary.csv
Untracked: code/annotating_fine-mapped_m6A_QTLs.Rmd
Untracked: code/check_double_strand.ipynb
Untracked: code/check_double_strand_v2.ipynb
Untracked: code/ctwas/
Untracked: code/figure/
Untracked: code/learn_gviz.Rmd
Untracked: code/learn_gviz.html
Untracked: code/learn_gviz.nb.html
Untracked: code/m6AQTL_finemapping.Rmd
Untracked: code/plot_genomic_tracks_gviz.ipynb
Untracked: code/summary_TWAS_coloc_m6A_2023.Rmd
Untracked: code/test_gviz.ipynb
Untracked: code/twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Untracked: data/.ipynb_checkpoints/
Untracked: data/ADCY7_gwas_input.tsv
Untracked: data/ADCY7_qtl_input.tsv
Untracked: data/Allergy_full_coloc.txt
Untracked: data/Asthma_full_coloc.txt
Untracked: data/CAD_full_coloc.txt
Untracked: data/Eosinophil_count_full_coloc.txt
Untracked: data/GSE125377_jointPeakReadCount.txt
Untracked: data/G_list.Rd
Untracked: data/HMGCR_ctwas_dat.Rd
Untracked: data/IBD_full_coloc.txt
Untracked: data/JointPeaks.bed
Untracked: data/Li2022_dsRNAs.xlsx
Untracked: data/Lupus_full_coloc.txt
Untracked: data/RA_full_coloc.txt
Untracked: data/TABLE1_hg19.txt
Untracked: data/TABLE1_hg19.txt.zip
Untracked: data/__MACOSX/
Untracked: data/coloc_blood_traits.csv
Untracked: data/crohns_disease_full_coloc.txt
Untracked: data/ctwas_m6a_joint_top_PIP.txt
Untracked: data/edit_sites_and_GE_neg_correlated.txt
Untracked: data/edit_sites_and_GE_pos_correlated.txt
Untracked: data/features
Untracked: data/human_EERs.csv
Untracked: data/human_EERs.txt
Untracked: data/lymph_full_coloc.txt
Untracked: data/m6A_TWAS_results.csv
Untracked: data/m6a_TWAS_genes.txt
Untracked: data/m6a_joint_calling_peaks.csv
Untracked: data/nasser_2021_ABC_IBD_genes.txt
Untracked: data/nat_sense_pairs.csv
Untracked: data/plt_full_coloc.txt
Untracked: data/rbc_full_coloc.txt
Untracked: data/rdw_full_coloc.txt
Untracked: data/reported_AS_targets_S1.txt
Untracked: data/reported_AS_wanowska.txt
Untracked: data/sig_coloc_results/
Untracked: data/test_locuscomparer.pdf
Untracked: data/ulcerative_colitis_full_coloc.txt
Untracked: data/wbc_full_coloc.txt
Untracked: data/zhao_silver_genes.csv
Untracked: output/.ipynb_checkpoints/
Untracked: output/HMGCR_gene_track_plot.pdf
Untracked: output/HMGCR_locus_plot.pdf
Untracked: output/IBD_DHX38_plot.pdf
Untracked: output/IBD_DHX38_plot_genetrack.pdf
Untracked: output/IBD_IP6K2_plot.pdf
Untracked: output/LDL_PMPCA_plot.pdf
Untracked: output/LDL_TMEM199_plot.pdf
Untracked: output/all_m6a_sites_with_cisNATs.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary_PP40.3.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary_PP40.5.csv
Untracked: output/all_m6a_sites_with_paired_cis_NATs.csv
Untracked: output/asthma_GPR183_plot.pdf
Untracked: output/asthma_GPR183_plot2.pdf
Untracked: output/asthma_GPR183_plot_genetrack.pdf
Untracked: output/asthma_GPR183_plot_genetrack2.pdf
Untracked: output/asthma_SLC25A11_plot_genetrack.pdf
Untracked: output/asthma_ZMAT2_plot.pdf
Untracked: output/asthma_ZMAT2_plot2.pdf
Untracked: output/asthma_ZMAT2_plot_genetrack.pdf
Untracked: output/asthma_ZMAT2_plot_genetrack2.pdf
Untracked: output/ctwas_locus_plot_output_data.Rd
Untracked: output/ctwas_m6a_joint_m6a_PIP_0.8.csv
Untracked: output/ctwas_m6a_joint_m6a_PIP_0.8.txt
Untracked: output/ctwas_m6a_joint_pip_above_0.6.RData
Untracked: output/eosinoph_MEPCE_plot.pdf
Untracked: output/eosinoph_PTK2B_plot.pdf
Untracked: output/eosinoph_PTK2B_plot_genetrack.pdf
Untracked: output/eosinoph_SH2D3C_plot.pdf
Untracked: output/eosinoph_SLC43A3_plot.pdf
Untracked: output/eosinoph_ZER1_plot.pdf
Untracked: output/eosinoph_ZER1_plot_genetrack.pdf
Untracked: output/fine_mapped_m6AQTLs_TWAS_genes_highPP4.rds
Untracked: output/gene_summary.csv
Untracked: output/immune_related_m6A_targets.csv
Untracked: output/lupus_MIR210HG_IRF7_plot.pdf
Untracked: output/lupus_MIR210HG_IRF7_plot_genetrack.pdf
Untracked: output/lupus_MIR210HG_plot.pdf
Untracked: output/lupus_MIR210HG_plot_genetrack.pdf
Untracked: output/lymph_ADCY7_BRD7_plot.pdf
Untracked: output/lymph_ADCY7_BRD7_plot_genetrack.pdf
Untracked: output/lymph_ADCY7_NOD2_plot.pdf
Untracked: output/lymph_ADCY7_NOD2_plot_genetrack.pdf
Untracked: output/lymph_ADCY7_plot.pdf
Untracked: output/lymph_BAZ1B_plot.pdf
Untracked: output/lymph_EPC1_plot.pdf
Untracked: output/lymph_LAMTOR4_plot.pdf
Untracked: output/lymph_MDM2_plot.pdf
Untracked: output/lymph_RANGAP1_plot.pdf
Untracked: output/lymph_TAP2_plot.pdf
Untracked: output/lymph_THEMIS2_plot.pdf
Untracked: output/lymph_ZKSCAN5_plot.pdf
Untracked: output/lymph_ZSCAN25_plot.pdf
Untracked: output/m6aQTL_dsRNAs_PPP2R3C_PRORP.pdf
Untracked: output/m6a_QTL_genes.csv
Untracked: output/m6a_genes_PIP_0.6_blood_immune.csv
Untracked: output/m6a_genes_PIP_0.6_blood_immune.txt
Untracked: output/m6a_peaks_nearby_dsRNAs.csv
Untracked: output/m6a_sites_near_all_dsRNAs_twas.csv
Untracked: output/m6a_sites_near_dsRNAs_coloc.csv
Untracked: output/m6a_sites_near_dsRNAs_twas.csv
Untracked: output/m6a_sites_near_dsRNAs_twas_summary.csv
Untracked: output/m6a_sites_overlapping_NAT_twas.csv
Untracked: output/m6a_sites_overlapping_dsRNAs_coloc.csv
Untracked: output/m6a_sites_overlapping_dsRNAs_twas.csv
Untracked: output/m6a_sites_overlapping_dsRegions.csv
Untracked: output/m6a_sites_overlapping_dsRegions_coloc.csv
Untracked: output/negatively_correlated_genes.txt
Untracked: output/platelet_AKAP8_plot.pdf
Untracked: output/platelet_C17orf62_plot.pdf
Untracked: output/platelet_GSDMD_plot.pdf
Untracked: output/platelet_PHF11_plot.pdf
Untracked: output/platelet_SLC25A11_plot.pdf
Untracked: output/platelet_TAOK1_plot.pdf
Untracked: output/platelet_THEMIS2_plot.pdf
Untracked: output/platelet_THEMIS2_plot_genetrack.pdf
Untracked: output/platelet_TRAF2_plot.pdf
Untracked: output/platelet_UBE2G2_plot.pdf
Untracked: output/platelet_ZSCAN25_plot.pdf
Untracked: output/postively_correlated_genes.txt
Untracked: output/rs1806261_RABEP1-NUP88_focused_locusview.pdf
Untracked: output/rs1806261_RABEP1-NUP88_locusview.pdf
Untracked: output/rs3177647_MAPKAPK5-AS1-MAPKAPK5_locusview.pdf
Untracked: output/rs3204541_DDX55-EIF2B1_locusview.pdf
Untracked: output/rs7184802_ADCY7-BRD7_locusview.pdf
Untracked: output/rs7184802_ADCY7_locuscompare.pdf
Untracked: output/twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Untracked: output/twas_genes_PP4_0.5_blood_traits_trackplots.pdf
Untracked: output/twas_m6a_sites_with_all_cisNATs.RDS
Untracked: output/twas_m6a_sites_with_cisNATs_range.RDS
Untracked: output/twas_m6a_sites_with_the_nearest_cisNAT.RDS
Untracked: output/wbc_RABEP1_NUP88_plot.pdf
Untracked: output/wbc_RABEP1_NUP88_plot_genetrack.pdf
Untracked: output/wbc_SLC9A3R1_plot.pdf
Untracked: twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Unstaged changes:
Deleted: analysis/learn_ctwas.Rmd
Modified: analysis/lymph_m6A_output_hg19.Rmd
Modified: analysis/m6A_switch_to_disease_h2g.Rmd
Modified: analysis/rbc_m6A_output_hg19.Rmd
Modified: analysis/wbc_m6A_output.Rmd
Modified: analysis/wbc_m6A_output_hg19.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/BMI_m6A_output_hg19.Rmd)
and HTML (docs/BMI_m6A_output_hg19.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d0f5634 | Jing Gu | 2023-08-25 | other traits |
Load ctwas results
# top 1 method
res <- impute_expr_z(z_snp, weight = weight, ld_R_dir = ld_R_dir,
method = NULL, outputdir = outputdir, outname = outname.e,
harmonize_z = T, harmonize_wgt = T, scale_by_ld_variance=F,
strand_ambig_action_z = "recover",
recover_strand_ambig_wgt = T
# lasso/elastic-net method
res <- impute_expr_z(z_snp, weight = weight, ld_R_dir = ld_R_dir,
method = NULL, outputdir = outputdir, outname = outname.e,
harmonize_z = T, harmonize_wgt = T, scale_by_ld_variance=F,
strand_ambig_action_z = "none",
recover_strand_ambig_wgt = FGWAS: UK Biobank GWAS summary statistics - European individuals
Weights: FUSION weights using top1, lasso, or elastic-net models were converted into PredictDB format and were not needed to do scaling when running ctwas.
Check convergence of parameters
cTWAS analysis on m6A alone
Joint analysis of expression, splicing and m6A
[1] "Check convergence for the lasso model when jointly analyzing expression, splicing and m6A:"
[1] "Table of group size before/after matching with UKBB SNPs:"
SNP eQTL sQTL m6AQTL
prior_group_size 9.324e+06 2005.0000 2191.000 918.0000
group_size 7.547e+06 1998.0000 2180.000 911.0000
percent_of_overlaps 8.094e-01 0.9965 0.995 0.9924
SNP eQTL sQTL m6AQTL
estimated_group_prior 2.791e-04 1.154e-02 2.617e-06 6.405e-03
estimated_group_prior_var 1.702e+01 1.382e+01 1.678e+01 1.249e+01
estimated_group_pve 1.066e-01 9.476e-04 2.847e-07 2.168e-04
attributable_group_pve 9.892e-01 8.790e-03 2.641e-06 2.011e-03$lasso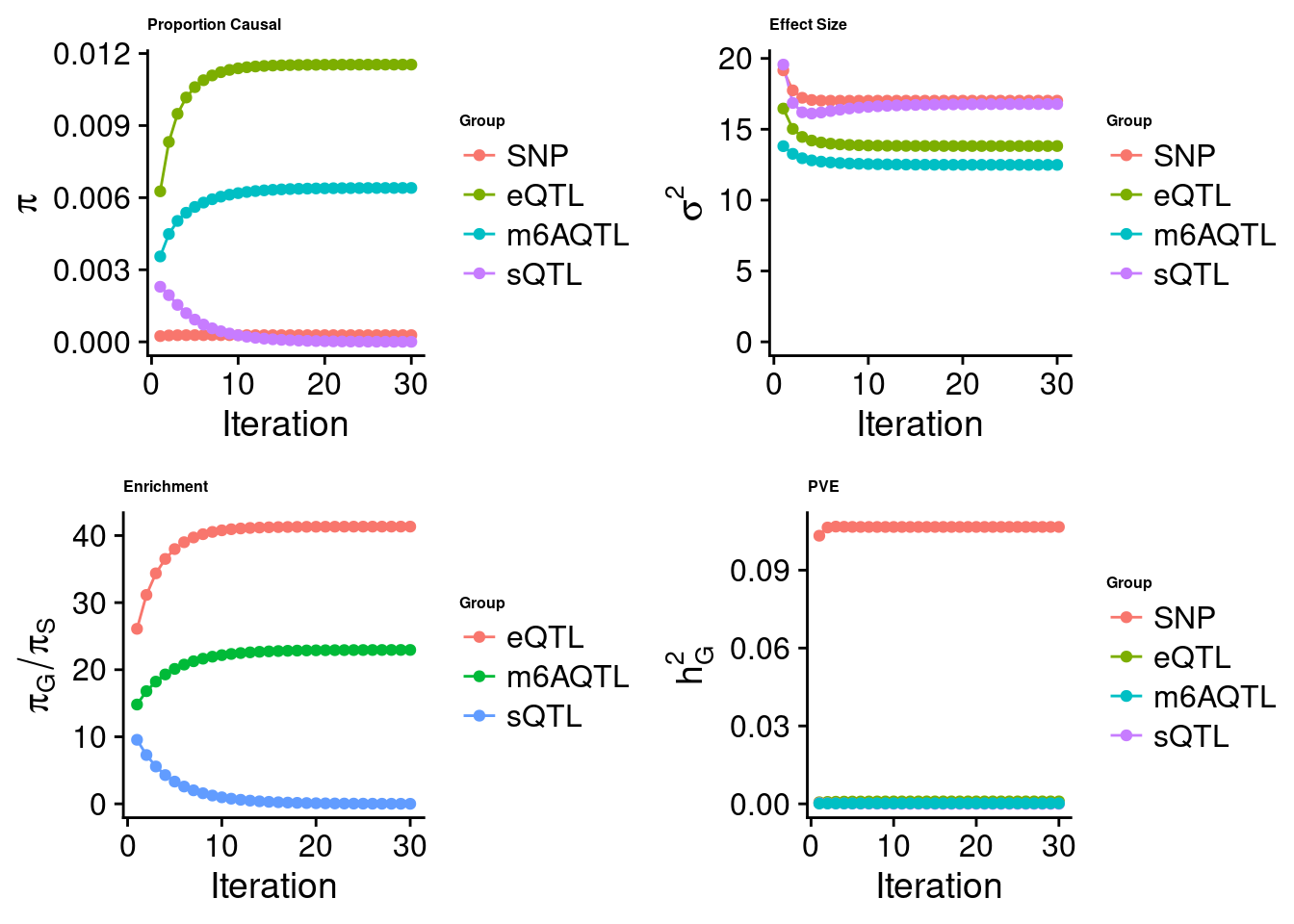
cTWAS results for individual analysis with m6A
top1 model
Summing up PIPs for m6A peaks located in the same gene
Top m6A PIPs by genes
cTWAS results for joint analysis using a lasso model
Top m6A modification pip
Top expression/splicing/m6A units
For m6A or splicing QTLs, they are assigned to the nearest genes (m6A needs to be confirmed with Kevin).
Top SNPs or genes with PIP > 0.6
$eQTL
genename susie_pip group region_tag
1987 AP1S1 0.7412 eQTL 7_62
1014 ENSG00000177236 0.7122 eQTL 11_1
731 STYXL1 0.6915 eQTL 7_48
$m6AQTL
genename susie_pip group region_tag
4542 AP3M2 0.631 m6AQTL 8_37
$sQTL
[1] genename susie_pip group region_tag
<0 rows> (or 0-length row.names)Top m6A modification pip
genename region_tag susie_pip z
1 AP3M2 8_37 0.6310 4.250
2 B4GALT5 20_30 0.4701 -3.275
3 FERMT3 11_36 0.3389 4.894
4 UCK1 9_70 0.2908 3.679
5 TRAPPC10 21_23 0.2477 -3.756
6 PIP4K2A 10_17 0.2297 3.476
7 RCN1 11_21 0.2219 -3.887
8 CYTH1 17_44 0.1977 -3.989
9 EXOG 3_28 0.1966 3.675
10 MXD4 4_4 0.1844 -4.033Summing up PIPs for m6A peaks located in the same gene
Top 10 m6A PIPs by genes
# A tibble: 818 × 2
genename total_susie_pip
<chr> <dbl>
1 AP3M2 0.631
2 B4GALT5 0.470
3 FERMT3 0.339
4 UCK1 0.291
5 TRAPPC10 0.248
6 PIP4K2A 0.230
7 RCN1 0.222
8 TBC1D4 0.198
9 CYTH1 0.198
10 EXOG 0.197
# ℹ 808 more rowsTop splicing PIPs
peak_id genename pos region_tag susie_pip z
1 chr8:128903244-128944711 MYC 128834403 8_84 0.0003342 -4.043
2 chr20:43516383-43530172 YWHAB 43514337 20_28 0.0002727 -4.436
3 chr20:43514527-43530172 YWHAB 43432189 20_28 0.0002515 4.414
4 chr5:96119784-96121492 ERAP1 96121524 5_57 0.0002473 4.565
5 chr17:7123848-7123923 ACADVL 7091650 17_8 0.0002457 -3.760
6 chr1:1770677-1779317 GNB1 1776269 1_1 0.0002218 -5.893
7 chr12:110934008-110937262 VPS29 110857694 12_67 0.0002153 -5.465
8 chr1:8930569-8931950 ENO1 8931463 1_6 0.0002129 4.232
9 chr3:16358739-16399439 RFTN1 16284694 3_12 0.0001778 -3.813
10 chr16:28845715-28845827 ATXN2L 28889486 16_23 0.0001660 -10.760Summing up PIPs for spliced introns located in the same gene
Top 10 splicing PIPs by genes
# A tibble: 10 × 2
genename total_susie_pip
<chr> <dbl>
1 YWHAB 0.000540
2 ATXN2L 0.000537
3 MYC 0.000344
4 ENO1 0.000300
5 GNB1 0.000290
6 ERAP1 0.000269
7 ACADVL 0.000257
8 VPS29 0.000215
9 DPH3 0.000203
10 OAS1 0.000183Top genes by combined PIP
genename combined_pip expression_pip splicing_pip m6A_pip
175 AP1S1 0.741 0.741208 0.000e+00 0.000000
872 ENSG00000177236 0.712 0.712174 0.000e+00 0.000000
2871 STYXL1 0.692 0.691497 1.519e-04 0.000000
178 AP3M2 0.653 0.021898 0.000e+00 0.630951
2176 PCYT1A 0.595 0.595136 0.000e+00 0.000000
2785 SNX11 0.548 0.546776 0.000e+00 0.001254
3286 YWHAZ 0.478 0.478461 0.000e+00 0.000000
278 B4GALT5 0.473 0.002616 0.000e+00 0.470118
1166 ENSG00000267080 0.458 0.457709 0.000e+00 0.000000
2593 RRN3 0.446 0.445719 0.000e+00 0.000000
3156 TYW5 0.432 0.431977 0.000e+00 0.000000
975 ENSG00000231365 0.375 0.375104 0.000e+00 0.000000
2641 SEC24C 0.359 0.358742 0.000e+00 0.000000
320 BPTF 0.357 0.356581 0.000e+00 0.000000
1327 FERMT3 0.341 0.002388 1.679e-07 0.338861
641 CTSW 0.338 0.337718 0.000e+00 0.000000
2438 RAPGEFL1 0.334 0.334152 0.000e+00 0.000000
337 BTN3A1 0.327 0.326972 0.000e+00 0.000000
3298 ZDBF2 0.326 0.326043 0.000e+00 0.000000
436 CCDC85B 0.324 0.323788 0.000e+00 0.000000
region_tag
175 7_62
872 11_1
2871 7_48
178 8_37
2176 3_121
2785 17_28
3286 8_69
278 20_30
1166 17_26
2593 16_15
3156 2_118
975 1_73
2641 10_49
320 17_39
1327 11_36
641 11_36
2438 17_23
337 6_20
3298 2_122
436 11_36Loading required package: gridWarning: replacing previous import 'utils::download.file' by
'restfulr::download.file' when loading 'rtracklayer'Locus plots for specific examples
R version 4.2.0 (2022-04-22)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=C LC_MONETARY=C LC_MESSAGES=C
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] biomaRt_2.52.0 Gviz_1.40.1 cowplot_1.1.1
[4] ggplot2_3.4.3 GenomicRanges_1.48.0 GenomeInfoDb_1.32.2
[7] IRanges_2.30.1 S4Vectors_0.34.0 BiocGenerics_0.42.0
[10] ctwas_0.1.38 dplyr_1.1.2 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] colorspace_2.1-0 deldir_1.0-6
[3] rjson_0.2.21 rprojroot_2.0.3
[5] biovizBase_1.44.0 htmlTable_2.4.0
[7] XVector_0.36.0 base64enc_0.1-3
[9] fs_1.6.3 dichromat_2.0-0.1
[11] rstudioapi_0.15.0 farver_2.1.1
[13] bit64_4.0.5 AnnotationDbi_1.58.0
[15] fansi_1.0.4 xml2_1.3.3
[17] codetools_0.2-18 logging_0.10-108
[19] cachem_1.0.8 knitr_1.39
[21] Formula_1.2-4 jsonlite_1.8.7
[23] Rsamtools_2.12.0 cluster_2.1.3
[25] dbplyr_2.3.3 png_0.1-7
[27] compiler_4.2.0 httr_1.4.7
[29] backports_1.4.1 lazyeval_0.2.2
[31] Matrix_1.6-1 fastmap_1.1.1
[33] cli_3.6.1 later_1.3.0
[35] htmltools_0.5.2 prettyunits_1.1.1
[37] tools_4.2.0 gtable_0.3.3
[39] glue_1.6.2 GenomeInfoDbData_1.2.8
[41] rappdirs_0.3.3 Rcpp_1.0.11
[43] Biobase_2.56.0 jquerylib_0.1.4
[45] vctrs_0.6.3 Biostrings_2.64.0
[47] rtracklayer_1.56.0 iterators_1.0.14
[49] xfun_0.30 stringr_1.5.0
[51] ps_1.7.0 lifecycle_1.0.3
[53] ensembldb_2.20.2 restfulr_0.0.14
[55] XML_3.99-0.14 getPass_0.2-2
[57] zlibbioc_1.42.0 scales_1.2.1
[59] BSgenome_1.64.0 VariantAnnotation_1.42.1
[61] ProtGenerics_1.28.0 hms_1.1.3
[63] promises_1.2.0.1 MatrixGenerics_1.8.0
[65] parallel_4.2.0 SummarizedExperiment_1.26.1
[67] AnnotationFilter_1.20.0 RColorBrewer_1.1-3
[69] yaml_2.3.5 curl_5.0.2
[71] memoise_2.0.1 gridExtra_2.3
[73] sass_0.4.1 rpart_4.1.16
[75] latticeExtra_0.6-30 stringi_1.7.12
[77] RSQLite_2.3.1 highr_0.9
[79] BiocIO_1.6.0 foreach_1.5.2
[81] checkmate_2.1.0 GenomicFeatures_1.48.4
[83] filelock_1.0.2 BiocParallel_1.30.3
[85] rlang_1.1.1 pkgconfig_2.0.3
[87] matrixStats_0.62.0 bitops_1.0-7
[89] evaluate_0.15 lattice_0.20-45
[91] htmlwidgets_1.5.4 GenomicAlignments_1.32.0
[93] labeling_0.4.2 bit_4.0.5
[95] processx_3.8.0 tidyselect_1.2.0
[97] magrittr_2.0.3 R6_2.5.1
[99] generics_0.1.3 Hmisc_5.1-0
[101] DelayedArray_0.22.0 DBI_1.1.3
[103] pgenlibr_0.3.6 pillar_1.9.0
[105] whisker_0.4 foreign_0.8-82
[107] withr_2.5.0 KEGGREST_1.36.2
[109] RCurl_1.98-1.7 nnet_7.3-17
[111] tibble_3.2.1 crayon_1.5.2
[113] interp_1.1-4 utf8_1.2.3
[115] BiocFileCache_2.4.0 rmarkdown_2.14
[117] jpeg_0.1-10 progress_1.2.2
[119] data.table_1.14.8 blob_1.2.4
[121] callr_3.7.3 git2r_0.30.1
[123] digest_0.6.33 httpuv_1.6.5
[125] munsell_0.5.0 bslib_0.3.1