cTWAS_analysis_for_lupus
Jing Gu
2023-08-17
Last updated: 2023-08-17
Checks: 6 1
Knit directory: m6A_in_disease_genetics/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230331) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/projects/m6A_in_disease_genetics/code/ctwas/ctwas_config_b37.R | code/ctwas/ctwas_config_b37.R |
| ~/projects/m6A_in_disease_genetics/code/ctwas/qiansheng/locus_plot.R | code/ctwas/qiansheng/locus_plot.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 06e2427. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: analysis/m6A_switch_to_disease_h2g.nb.html
Ignored: data/plots/
Untracked files:
Untracked: HMGCR_locus_gene_tracks.pdf
Untracked: Rplots.pdf
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/IBD_E_S_m6A.Rmd
Untracked: analysis/IBD_E_S_m6A_output.Rmd
Untracked: analysis/LDL_E_S_m6A.Rmd
Untracked: analysis/LDL_m6A_output.Rmd
Untracked: analysis/RA_m6A_output.Rmd
Untracked: analysis/WhiteBlood_WholeBlood_E_M.Rmd
Untracked: analysis/identify_m6A_mechanisms_with_finemapping.Rmd
Untracked: analysis/lymph_m6A_output.Rmd
Untracked: analysis/pre_weights_m6AQTL.txt
Untracked: analysis/rbc_E_S_m6A_output.Rmd
Untracked: analysis/rbc_m6A_output.Rmd
Untracked: analysis/wbc_E_S_m6A_output.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/all_m6a_sites_with_paired_cisNATs_summary.csv
Untracked: code/annotating_fine-mapped_m6A_QTLs.Rmd
Untracked: code/check_double_strand.ipynb
Untracked: code/check_double_strand_v2.ipynb
Untracked: code/ctwas/
Untracked: code/figure/
Untracked: code/learn_gviz.Rmd
Untracked: code/learn_gviz.html
Untracked: code/learn_gviz.nb.html
Untracked: code/m6AQTL_finemapping.Rmd
Untracked: code/summary_TWAS_coloc_m6A_2023.Rmd
Untracked: code/test_gviz.ipynb
Untracked: code/twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Untracked: data/.ipynb_checkpoints/
Untracked: data/ADCY7_gwas_input.tsv
Untracked: data/ADCY7_qtl_input.tsv
Untracked: data/Allergy_full_coloc.txt
Untracked: data/Asthma_full_coloc.txt
Untracked: data/CAD_full_coloc.txt
Untracked: data/Eosinophil_count_full_coloc.txt
Untracked: data/GSE125377_jointPeakReadCount.txt
Untracked: data/HMGCR_ctwas_dat.Rd
Untracked: data/IBD_full_coloc.txt
Untracked: data/JointPeaks.bed
Untracked: data/Li2022_dsRNAs.xlsx
Untracked: data/Lupus_full_coloc.txt
Untracked: data/RA_full_coloc.txt
Untracked: data/TABLE1_hg19.txt
Untracked: data/TABLE1_hg19.txt.zip
Untracked: data/__MACOSX/
Untracked: data/coloc_blood_traits.csv
Untracked: data/crohns_disease_full_coloc.txt
Untracked: data/edit_sites_and_GE_neg_correlated.txt
Untracked: data/edit_sites_and_GE_pos_correlated.txt
Untracked: data/features
Untracked: data/human_EERs.csv
Untracked: data/human_EERs.txt
Untracked: data/lymph_full_coloc.txt
Untracked: data/m6A_TWAS_results.csv
Untracked: data/m6a_TWAS_genes.txt
Untracked: data/m6a_joint_calling_peaks.csv
Untracked: data/nasser_2021_ABC_IBD_genes.txt
Untracked: data/nat_sense_pairs.csv
Untracked: data/plt_full_coloc.txt
Untracked: data/rbc_full_coloc.txt
Untracked: data/rdw_full_coloc.txt
Untracked: data/reported_AS_targets_S1.txt
Untracked: data/reported_AS_wanowska.txt
Untracked: data/sig_coloc_results/
Untracked: data/test_locuscomparer.pdf
Untracked: data/ulcerative_colitis_full_coloc.txt
Untracked: data/wbc_full_coloc.txt
Untracked: data/zhao_silver_genes.csv
Untracked: output/.ipynb_checkpoints/
Untracked: output/HMGCR_gene_track_plot.pdf
Untracked: output/HMGCR_locus_plot.pdf
Untracked: output/all_m6a_sites_with_cisNATs.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary_PP40.3.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary_PP40.5.csv
Untracked: output/all_m6a_sites_with_paired_cis_NATs.csv
Untracked: output/fine_mapped_m6AQTLs_TWAS_genes_highPP4.rds
Untracked: output/gene_summary.csv
Untracked: output/immune_related_m6A_targets.csv
Untracked: output/m6aQTL_dsRNAs_PPP2R3C_PRORP.pdf
Untracked: output/m6a_peaks_nearby_dsRNAs.csv
Untracked: output/m6a_sites_near_all_dsRNAs_twas.csv
Untracked: output/m6a_sites_near_dsRNAs_coloc.csv
Untracked: output/m6a_sites_near_dsRNAs_twas.csv
Untracked: output/m6a_sites_near_dsRNAs_twas_summary.csv
Untracked: output/m6a_sites_overlapping_NAT_twas.csv
Untracked: output/m6a_sites_overlapping_dsRNAs_coloc.csv
Untracked: output/m6a_sites_overlapping_dsRNAs_twas.csv
Untracked: output/m6a_sites_overlapping_dsRegions.csv
Untracked: output/m6a_sites_overlapping_dsRegions_coloc.csv
Untracked: output/negatively_correlated_genes.txt
Untracked: output/postively_correlated_genes.txt
Untracked: output/rs1806261_RABEP1-NUP88_focused_locusview.pdf
Untracked: output/rs1806261_RABEP1-NUP88_locusview.pdf
Untracked: output/rs3177647_MAPKAPK5-AS1-MAPKAPK5_locusview.pdf
Untracked: output/rs3204541_DDX55-EIF2B1_locusview.pdf
Untracked: output/rs7184802_ADCY7-BRD7_locusview.pdf
Untracked: output/rs7184802_ADCY7_locuscompare.pdf
Untracked: output/twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Untracked: output/twas_genes_PP4_0.5_blood_traits_trackplots.pdf
Untracked: output/twas_m6a_sites_with_all_cisNATs.RDS
Untracked: output/twas_m6a_sites_with_cisNATs_range.RDS
Untracked: output/twas_m6a_sites_with_the_nearest_cisNAT.RDS
Untracked: twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Unstaged changes:
Modified: analysis/lymph_m6A_output_hg19.Rmd
Modified: analysis/m6A_switch_to_disease_h2g.Rmd
Modified: analysis/wbc_m6A_output.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/lupus_m6A_output_hg19.Rmd)
and HTML (docs/lupus_m6A_output_hg19.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 06e2427 | Jing Gu | 2023-08-17 | added lupus |
Load ctwas results
# top 1 method
res <- impute_expr_z(z_snp, weight = weight, ld_R_dir = ld_R_dir,
method = NULL, outputdir = outputdir, outname = outname.e,
harmonize_z = T, harmonize_wgt = T, scale_by_ld_variance=F,
strand_ambig_action_z = "recover",
recover_strand_ambig_wgt = T
# lasso/elastic-net method
res <- impute_expr_z(z_snp, weight = weight, ld_R_dir = ld_R_dir,
method = NULL, outputdir = outputdir, outname = outname.e,
harmonize_z = T, harmonize_wgt = T, scale_by_ld_variance=F,
strand_ambig_action_z = "none",
recover_strand_ambig_wgt = FGWAS: UK Biobank GWAS summary statistics - European individuals
Weights: FUSION weights using top1, lasso, or elastic-net models were converted into PredictDB format and were not needed to do scaling when running ctwas.
Check convergence of parameters
cTWAS analysis on m6A alone
[1] "Check convergence for the top1 model:"
[1] "Table of group size:"
SNP gene
6731160 801
SNP gene
estimated_group_prior 0.0002642 0.0010266
estimated_group_prior_var 5.2751345 1.8792046
estimated_group_pve 0.6574782 0.0001083
attributable_group_pve 0.9998353 0.0001647$top1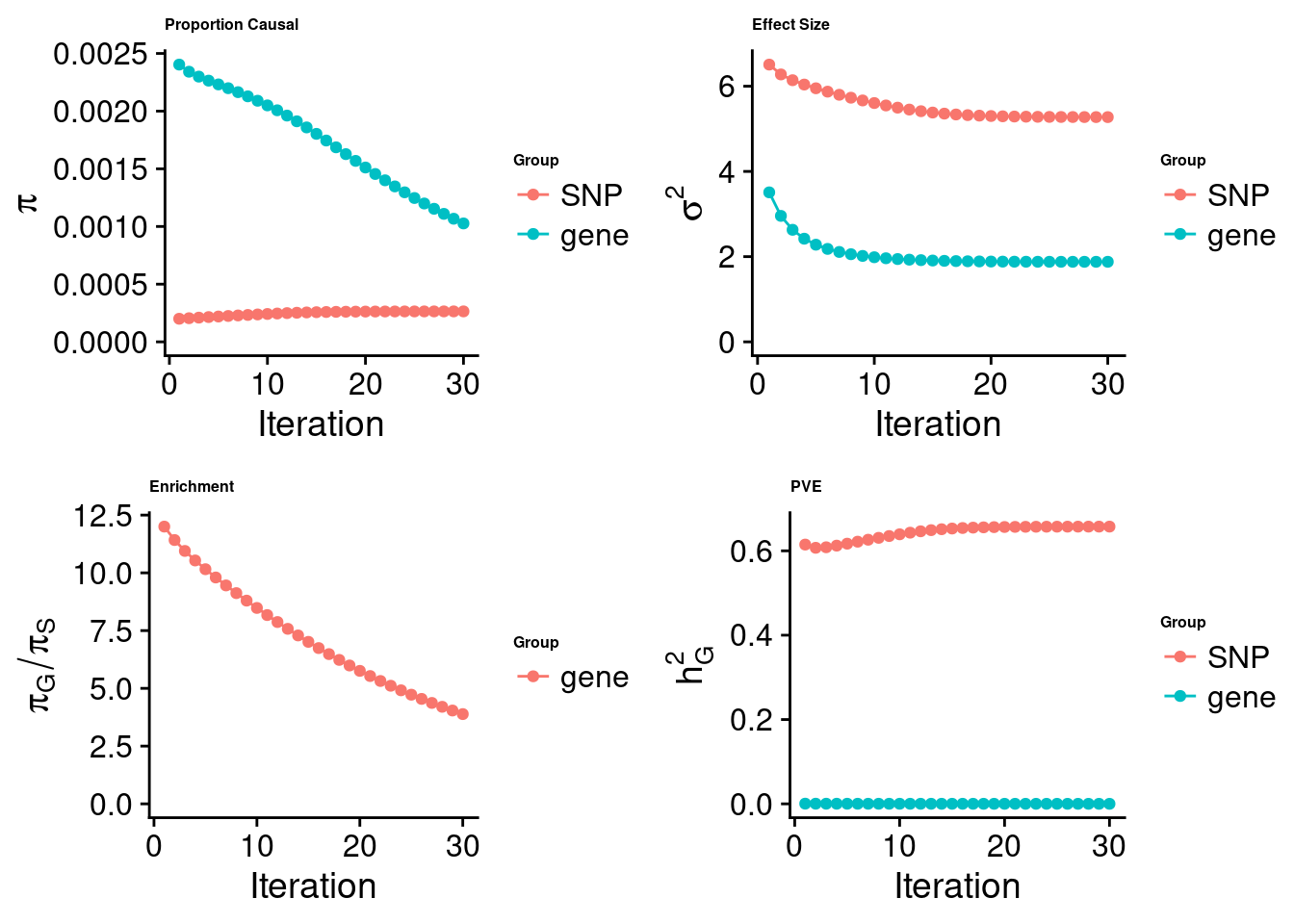
Joint analysis of expression, splicing and m6A
[1] "Check convergence for the lasso model when jointly analyzing expression, splicing and m6A:"
[1] "Table of group size before/after matching with UKBB SNPs:"
SNP eQTL sQTL m6AQTL
prior_group_size 9.324e+06 2005.0000 2191.0000 918.0000
group_size 6.731e+06 1984.0000 2149.0000 902.0000
percent_of_overlaps 7.219e-01 0.9895 0.9808 0.9826
SNP eQTL sQTL m6AQTL
estimated_group_prior 0.0002261 0.017777 0.002642 0.004184
estimated_group_prior_var 6.3614278 4.000501 3.878062 22.909135
estimated_group_pve 0.6787153 0.009889 0.001544 0.006061
attributable_group_pve 0.9748732 0.014205 0.002217 0.008705$lasso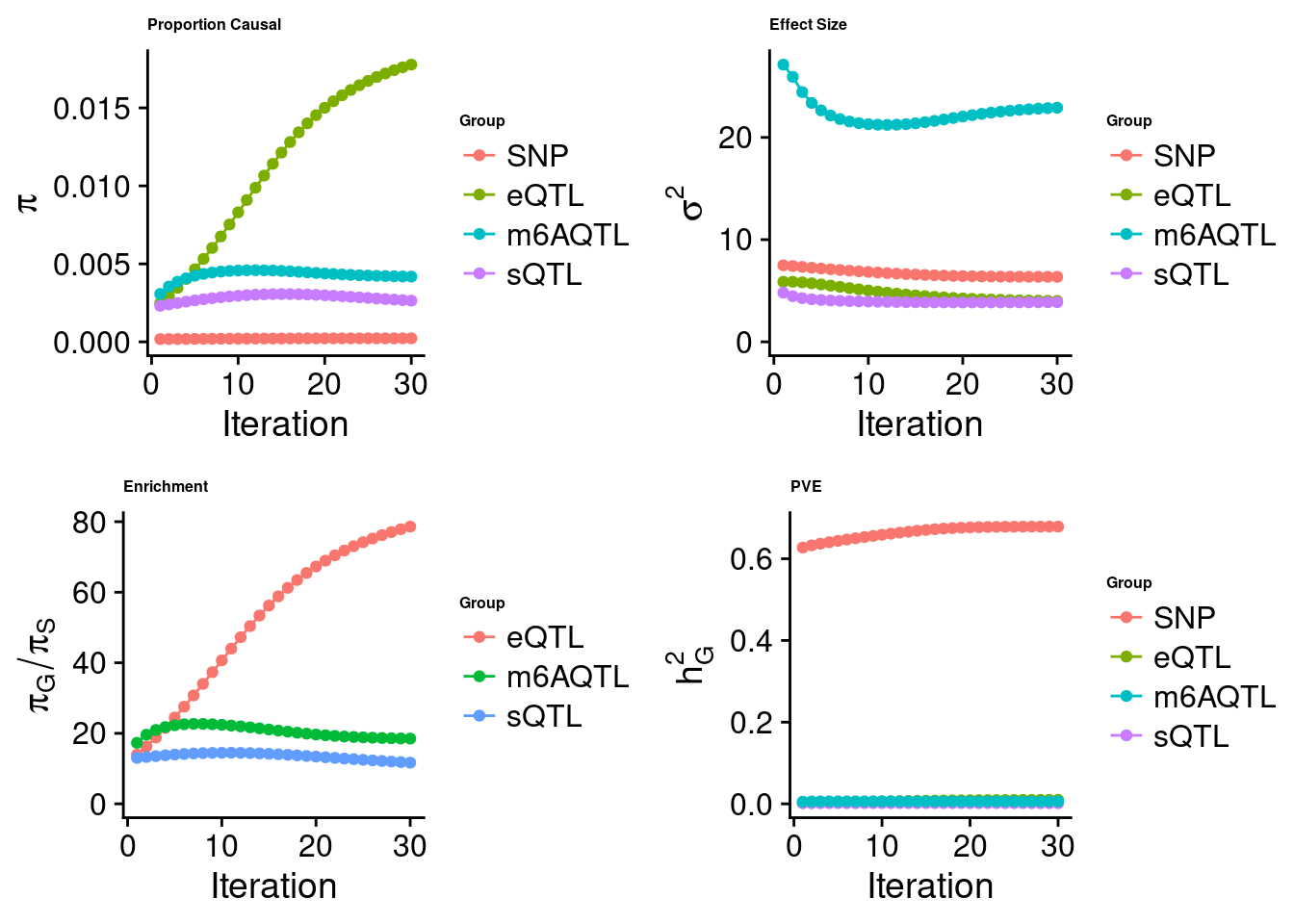
cTWAS results for individual analysis with m6A
top1 model
[1] genename region_tag susie_pip z
<0 rows> (or 0-length row.names)Summing up PIPs for m6A peaks located in the same gene
Top m6A PIPs by genes
# A tibble: 0 × 2
# ℹ 2 variables: genename <chr>, total_susie_pip <dbl>cTWAS results for joint analysis using a lasso model
Top m6A modification pip
Top expression/splicing/m6A units
For m6A or splicing QTLs, they are assigned to the nearest genes (m6A needs to be confirmed with Kevin).
Top SNPs or genes with PIP > 0.6
$eQTL
genename susie_pip group region_tag
1970 JADE2 0.6801 eQTL 5_80
$m6AQTL
genename susie_pip group region_tag
5032 MIR210HG 0.906 m6AQTL 11_1
$sQTL
[1] genename susie_pip group region_tag
<0 rows> (or 0-length row.names)Top m6A modification pip
genename region_tag susie_pip z
1 MIR210HG 11_1 0.90599 6.8092
2 IRF5 7_80 0.16925 0.6668
3 ICOSLG 21_23 0.09542 -3.1780
4 SH2D3C 9_66 0.09282 3.0549
5 IRF3 19_34 0.07407 -3.6731
6 SCIMP 17_5 0.07083 2.9299
7 PSMB1 6_112 0.06819 -2.8428
8 MKRN2 3_9 0.06440 2.8379
9 SNRNP25 16_1 0.06333 -2.8226
10 DENND3 8_92 0.06004 2.5839Summing up PIPs for m6A peaks located in the same gene
Top 10 m6A PIPs by genes
# A tibble: 809 × 2
genename total_susie_pip
<chr> <dbl>
1 MIR210HG 0.906
2 IRF5 0.169
3 SH2D3C 0.131
4 ICOSLG 0.101
5 IRF3 0.0741
6 SCIMP 0.0708
7 PSMB1 0.0682
8 MKRN2 0.0644
9 SNRNP25 0.0633
10 DENND3 0.0600
# ℹ 799 more rowsTop splicing PIPs
peak_id genename pos region_tag susie_pip z
1 chr22:42000157-42001876 DESI1 41914593 22_17 0.21438 3.351
2 chr20:44750537-44750690 CD40 44674743 20_28 0.15765 -3.713
3 chr14:24902238-24903306 KHNYN 24810413 14_4 0.14932 3.663
4 chr1:150240527-150241098 APH1A 150210120 1_75 0.14692 -3.263
5 chr19:50166515-50167931 IRF3 50093572 19_34 0.12050 -4.125
6 chr6:6589103-6625159 LY86 6521970 6_6 0.11113 3.131
7 chr12:6878840-6879010 PTMS 6870549 12_7 0.10886 -2.963
8 chr19:50168103-50168929 IRF3 50168871 19_34 0.09584 4.026
9 chr3:16305736-16306276 DPH3 16395668 3_12 0.09063 -2.866
10 chr20:5090091-5092142 TMEM230 5055138 20_4 0.08408 -3.286Summing up PIPs for spliced introns located in the same gene
Top 10 splicing PIPs by genes
# A tibble: 10 × 2
genename total_susie_pip
<chr> <dbl>
1 CD40 0.309
2 IFI44L 0.307
3 WARS1 0.278
4 IRF3 0.223
5 DESI1 0.214
6 TMEM230 0.206
7 NADSYN1 0.177
8 LBP 0.163
9 PDCD2 0.163
10 MGST3 0.163Top genes by combined PIP
genename combined_pip expression_pip splicing_pip m6A_pip
1838 MIR210HG 0.906 0.00000 0.00000 0.905991
1602 JADE2 0.704 0.68005 0.02365 0.000000
1098 ENSG00000257073 0.488 0.48833 0.00000 0.000000
304 BIK 0.481 0.48108 0.00000 0.000000
3215 WARS1 0.468 0.19047 0.27791 0.000000
1315 FCRLA 0.419 0.37282 0.04602 0.000000
269 AZI2 0.378 0.37755 0.00000 0.000000
2960 TMA16 0.371 0.37074 0.00000 0.000000
760 DRAM2 0.360 0.24891 0.11092 0.000000
2763 SNX11 0.351 0.34579 0.00000 0.004929
1030 ENSG00000244625 0.348 0.34797 0.00000 0.000000
1954 NADSYN1 0.342 0.16519 0.17699 0.000000
694 DEF6 0.332 0.33198 0.00000 0.000000
1542 IFI44L 0.320 0.00000 0.30674 0.013162
467 CD40 0.309 0.00000 0.30888 0.000000
640 CTTNBP2NL 0.305 0.30490 0.00000 0.000000
2215 PITPNC1 0.303 0.30273 0.00000 0.000000
181 AP5B1 0.298 0.29767 0.00000 0.000000
2262 POLR1E 0.298 0.25222 0.01010 0.035767
1584 IRF3 0.297 0.00000 0.22294 0.074067
2554 RPS19BP1 0.297 0.29678 0.00000 0.000000
2965 TMCO4 0.296 0.29551 0.00000 0.000000
2006 NFRKB 0.288 0.28830 0.00000 0.000000
705 DESI1 0.285 0.07062 0.21438 0.000000
491 CDK11A 0.279 0.27856 0.00000 0.000000
151 ANKDD1A 0.265 0.26522 0.00000 0.000000
187 APIP 0.265 0.22475 0.04049 0.000000
1585 IRF5 0.265 0.01898 0.07662 0.169246
1830 MIB2 0.262 0.26245 0.00000 0.000000
719 DHTKD1 0.261 0.26086 0.00000 0.000000
2999 TMEM44 0.261 0.26106 0.00000 0.000000
2863 SWI5 0.259 0.25876 0.00000 0.000000
1428 GSDMD 0.255 0.19767 0.00000 0.057483
1827 MGST3 0.255 0.09258 0.16291 0.000000
824 ELL2 0.253 0.25318 0.00000 0.000000
3204 VPS16 0.253 0.25326 0.00000 0.000000
1682 LHPP 0.248 0.24766 0.00000 0.000000
210 ARL5B 0.247 0.24684 0.00000 0.000000
903 ENSG00000211891 0.247 0.24720 0.00000 0.000000
2370 PTPRA 0.242 0.20415 0.02904 0.008818
region_tag
1838 11_1
1602 5_80
1098 17_42
304 22_18
3215 14_52
1315 1_81
269 3_21
2960 4_105
760 1_69
2763 17_28
1030 22_8
1954 11_40
694 6_28
1542 1_48
467 20_28
640 1_69
2215 17_39
181 11_36
2262 9_28
1584 19_34
2554 22_16
2965 1_13
2006 11_80
705 22_17
491 1_1
151 15_30
187 11_23
1585 7_80
1830 1_1
719 10_10
2999 3_119
2863 9_66
1428 8_94
1827 1_83
824 5_56
3204 20_3
1682 10_78
210 10_15
903 14_55
2370 20_3Loading required package: gridWarning: replacing previous import 'utils::download.file' by
'restfulr::download.file' when loading 'rtracklayer'Locus plots for specific examples
Some literature that relates MIR210HG to m6A modification
genename combined_pip expression_pip splicing_pip m6A_pip region_tag
1838 MIR210HG 0.906 0 0 0.906 11_1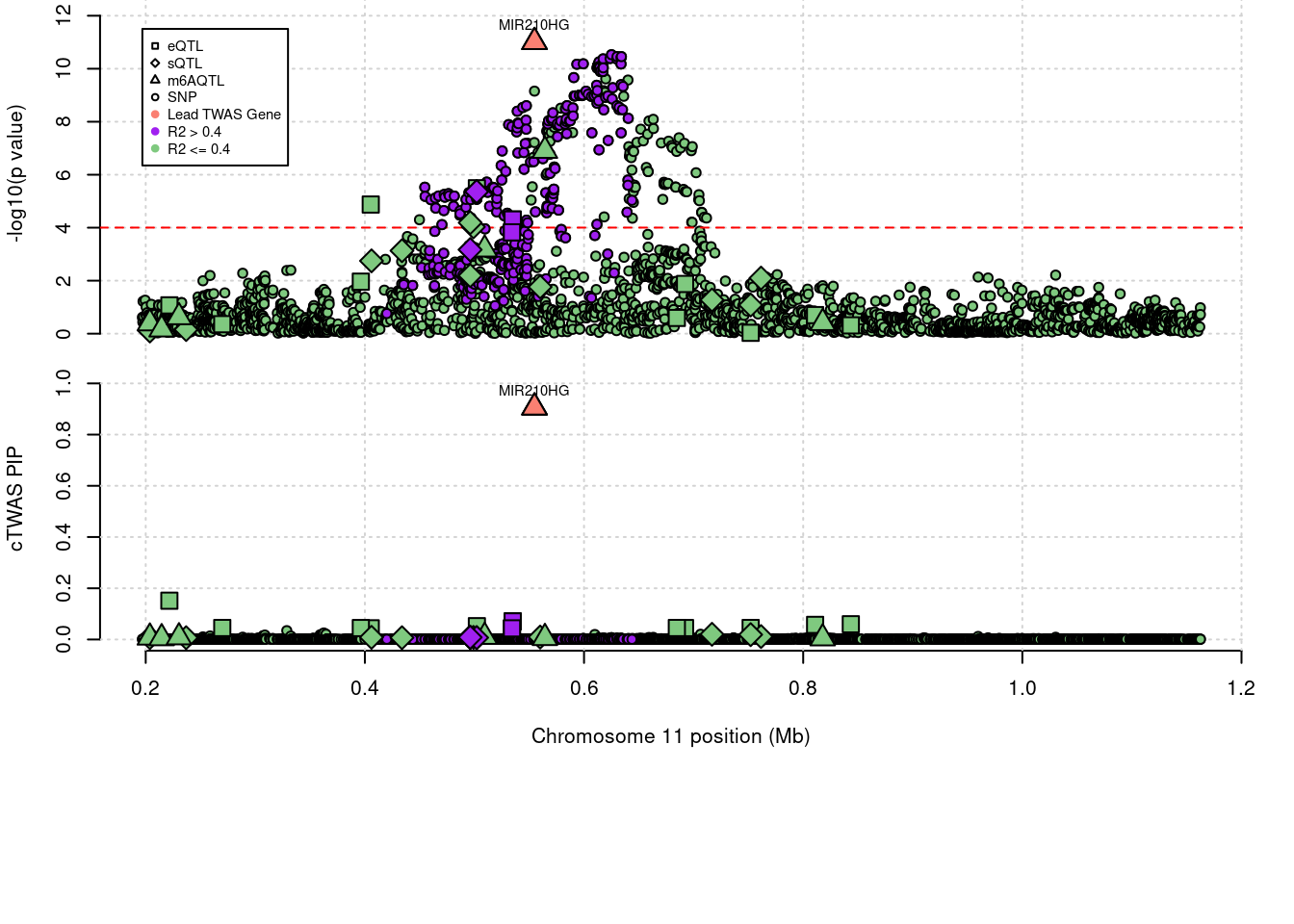
R version 4.2.0 (2022-04-22)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=C LC_MONETARY=C LC_MESSAGES=C
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] biomaRt_2.52.0 Gviz_1.40.1 cowplot_1.1.1
[4] ggplot2_3.4.3 GenomicRanges_1.48.0 GenomeInfoDb_1.32.2
[7] IRanges_2.30.1 S4Vectors_0.34.0 BiocGenerics_0.42.0
[10] ctwas_0.1.38 dplyr_1.1.2 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] colorspace_2.1-0 deldir_1.0-6
[3] rjson_0.2.21 rprojroot_2.0.3
[5] biovizBase_1.44.0 htmlTable_2.4.0
[7] XVector_0.36.0 base64enc_0.1-3
[9] fs_1.6.3 dichromat_2.0-0.1
[11] rstudioapi_0.15.0 farver_2.1.1
[13] bit64_4.0.5 AnnotationDbi_1.58.0
[15] fansi_1.0.4 xml2_1.3.3
[17] codetools_0.2-18 logging_0.10-108
[19] cachem_1.0.8 knitr_1.39
[21] Formula_1.2-4 jsonlite_1.8.7
[23] Rsamtools_2.12.0 cluster_2.1.3
[25] dbplyr_2.3.3 png_0.1-7
[27] compiler_4.2.0 httr_1.4.6
[29] backports_1.4.1 lazyeval_0.2.2
[31] Matrix_1.6-1 fastmap_1.1.1
[33] cli_3.6.1 later_1.3.0
[35] htmltools_0.5.2 prettyunits_1.1.1
[37] tools_4.2.0 gtable_0.3.3
[39] glue_1.6.2 GenomeInfoDbData_1.2.8
[41] rappdirs_0.3.3 Rcpp_1.0.11
[43] Biobase_2.56.0 jquerylib_0.1.4
[45] vctrs_0.6.3 Biostrings_2.64.0
[47] rtracklayer_1.56.0 iterators_1.0.14
[49] xfun_0.30 stringr_1.5.0
[51] ps_1.7.0 lifecycle_1.0.3
[53] ensembldb_2.20.2 restfulr_0.0.14
[55] XML_3.99-0.14 getPass_0.2-2
[57] zlibbioc_1.42.0 scales_1.2.1
[59] BSgenome_1.64.0 VariantAnnotation_1.42.1
[61] ProtGenerics_1.28.0 hms_1.1.3
[63] promises_1.2.0.1 MatrixGenerics_1.8.0
[65] parallel_4.2.0 SummarizedExperiment_1.26.1
[67] AnnotationFilter_1.20.0 RColorBrewer_1.1-3
[69] yaml_2.3.5 curl_5.0.2
[71] memoise_2.0.1 gridExtra_2.3
[73] sass_0.4.1 rpart_4.1.16
[75] latticeExtra_0.6-30 stringi_1.7.12
[77] RSQLite_2.3.1 highr_0.9
[79] BiocIO_1.6.0 foreach_1.5.2
[81] checkmate_2.1.0 GenomicFeatures_1.48.4
[83] filelock_1.0.2 BiocParallel_1.30.3
[85] rlang_1.1.1 pkgconfig_2.0.3
[87] matrixStats_0.62.0 bitops_1.0-7
[89] evaluate_0.15 lattice_0.20-45
[91] htmlwidgets_1.5.4 GenomicAlignments_1.32.0
[93] labeling_0.4.2 bit_4.0.5
[95] processx_3.8.0 tidyselect_1.2.0
[97] magrittr_2.0.3 R6_2.5.1
[99] generics_0.1.3 Hmisc_5.1-0
[101] DelayedArray_0.22.0 DBI_1.1.3
[103] pgenlibr_0.3.6 pillar_1.9.0
[105] whisker_0.4 foreign_0.8-82
[107] withr_2.5.0 KEGGREST_1.36.2
[109] RCurl_1.98-1.7 nnet_7.3-17
[111] tibble_3.2.1 crayon_1.5.2
[113] interp_1.1-4 utf8_1.2.3
[115] BiocFileCache_2.4.0 rmarkdown_2.14
[117] jpeg_0.1-10 progress_1.2.2
[119] data.table_1.14.8 blob_1.2.4
[121] callr_3.7.3 git2r_0.30.1
[123] digest_0.6.33 httpuv_1.6.5
[125] munsell_0.5.0 bslib_0.3.1