cTWAS_analysis_for_lymph
Jing Gu
2023-08-16
Last updated: 2023-08-16
Checks: 6 1
Knit directory: m6A_in_disease_genetics/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230331) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/projects/m6A_in_disease_genetics/code/ctwas/ctwas_config_b37.R | code/ctwas/ctwas_config_b37.R |
| ~/projects/m6A_in_disease_genetics/code/ctwas/qiansheng/locus_plot.R | code/ctwas/qiansheng/locus_plot.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 08eaf44. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: analysis/m6A_switch_to_disease_h2g.nb.html
Ignored: data/plots/
Untracked files:
Untracked: HMGCR_locus_gene_tracks.pdf
Untracked: Rplots.pdf
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/IBD_E_S_m6A.Rmd
Untracked: analysis/IBD_E_S_m6A_output.Rmd
Untracked: analysis/LDL_E_S_m6A.Rmd
Untracked: analysis/LDL_m6A_output.Rmd
Untracked: analysis/RA_m6A_output.Rmd
Untracked: analysis/WhiteBlood_WholeBlood_E_M.Rmd
Untracked: analysis/identify_m6A_mechanisms_with_finemapping.Rmd
Untracked: analysis/lymph_m6A_output.Rmd
Untracked: analysis/pre_weights_m6AQTL.txt
Untracked: analysis/rbc_E_S_m6A_output.Rmd
Untracked: analysis/rbc_m6A_output.Rmd
Untracked: analysis/rbc_m6A_output_hg19.Rmd
Untracked: analysis/wbc_E_S_m6A_output.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/all_m6a_sites_with_paired_cisNATs_summary.csv
Untracked: code/check_double_strand.ipynb
Untracked: code/check_double_strand_v2.ipynb
Untracked: code/ctwas/
Untracked: code/figure/
Untracked: code/learn_gviz.Rmd
Untracked: code/learn_gviz.html
Untracked: code/learn_gviz.nb.html
Untracked: code/m6AQTL_finemapping.Rmd
Untracked: code/summary_TWAS_coloc_m6A_2023.Rmd
Untracked: code/test_gviz.ipynb
Untracked: code/twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Untracked: data/.ipynb_checkpoints/
Untracked: data/ADCY7_gwas_input.tsv
Untracked: data/ADCY7_qtl_input.tsv
Untracked: data/Allergy_full_coloc.txt
Untracked: data/Asthma_full_coloc.txt
Untracked: data/CAD_full_coloc.txt
Untracked: data/Eosinophil_count_full_coloc.txt
Untracked: data/GSE125377_jointPeakReadCount.txt
Untracked: data/HMGCR_ctwas_dat.Rd
Untracked: data/IBD_full_coloc.txt
Untracked: data/JointPeaks.bed
Untracked: data/Li2022_dsRNAs.xlsx
Untracked: data/Lupus_full_coloc.txt
Untracked: data/RA_full_coloc.txt
Untracked: data/TABLE1_hg19.txt
Untracked: data/TABLE1_hg19.txt.zip
Untracked: data/__MACOSX/
Untracked: data/coloc_blood_traits.csv
Untracked: data/crohns_disease_full_coloc.txt
Untracked: data/edit_sites_and_GE_neg_correlated.txt
Untracked: data/edit_sites_and_GE_pos_correlated.txt
Untracked: data/features
Untracked: data/human_EERs.csv
Untracked: data/human_EERs.txt
Untracked: data/lymph_full_coloc.txt
Untracked: data/m6A_TWAS_results.csv
Untracked: data/m6a_TWAS_genes.txt
Untracked: data/m6a_joint_calling_peaks.csv
Untracked: data/nasser_2021_ABC_IBD_genes.txt
Untracked: data/nat_sense_pairs.csv
Untracked: data/plt_full_coloc.txt
Untracked: data/rbc_full_coloc.txt
Untracked: data/rdw_full_coloc.txt
Untracked: data/reported_AS_targets_S1.txt
Untracked: data/reported_AS_wanowska.txt
Untracked: data/sig_coloc_results/
Untracked: data/test_locuscomparer.pdf
Untracked: data/ulcerative_colitis_full_coloc.txt
Untracked: data/wbc_full_coloc.txt
Untracked: data/zhao_silver_genes.csv
Untracked: output/.ipynb_checkpoints/
Untracked: output/HMGCR_gene_track_plot.pdf
Untracked: output/HMGCR_locus_plot.pdf
Untracked: output/all_m6a_sites_with_cisNATs.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary_PP40.3.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary_PP40.5.csv
Untracked: output/all_m6a_sites_with_paired_cis_NATs.csv
Untracked: output/fine_mapped_m6AQTLs_TWAS_genes_highPP4.rds
Untracked: output/gene_summary.csv
Untracked: output/immune_related_m6A_targets.csv
Untracked: output/m6aQTL_dsRNAs_PPP2R3C_PRORP.pdf
Untracked: output/m6a_peaks_nearby_dsRNAs.csv
Untracked: output/m6a_sites_near_all_dsRNAs_twas.csv
Untracked: output/m6a_sites_near_dsRNAs_coloc.csv
Untracked: output/m6a_sites_near_dsRNAs_twas.csv
Untracked: output/m6a_sites_near_dsRNAs_twas_summary.csv
Untracked: output/m6a_sites_overlapping_NAT_twas.csv
Untracked: output/m6a_sites_overlapping_dsRNAs_coloc.csv
Untracked: output/m6a_sites_overlapping_dsRNAs_twas.csv
Untracked: output/m6a_sites_overlapping_dsRegions.csv
Untracked: output/m6a_sites_overlapping_dsRegions_coloc.csv
Untracked: output/negatively_correlated_genes.txt
Untracked: output/postively_correlated_genes.txt
Untracked: output/rs1806261_RABEP1-NUP88_focused_locusview.pdf
Untracked: output/rs1806261_RABEP1-NUP88_locusview.pdf
Untracked: output/rs3177647_MAPKAPK5-AS1-MAPKAPK5_locusview.pdf
Untracked: output/rs3204541_DDX55-EIF2B1_locusview.pdf
Untracked: output/rs7184802_ADCY7-BRD7_locusview.pdf
Untracked: output/rs7184802_ADCY7_locuscompare.pdf
Untracked: output/twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Untracked: output/twas_genes_PP4_0.5_blood_traits_trackplots.pdf
Untracked: output/twas_m6a_sites_with_all_cisNATs.RDS
Untracked: output/twas_m6a_sites_with_cisNATs_range.RDS
Untracked: output/twas_m6a_sites_with_the_nearest_cisNAT.RDS
Untracked: twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Unstaged changes:
Modified: analysis/m6A_switch_to_disease_h2g.Rmd
Modified: analysis/wbc_m6A_output.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/lymph_m6A_output_hg19.Rmd)
and HTML (docs/lymph_m6A_output_hg19.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 08eaf44 | Jing Gu | 2023-08-16 | run ctwas for multiple traits |
Load ctwas results
# top 1 method
res <- impute_expr_z(z_snp, weight = weight, ld_R_dir = ld_R_dir,
method = NULL, outputdir = outputdir, outname = outname.e,
harmonize_z = T, harmonize_wgt = T, scale_by_ld_variance=F,
strand_ambig_action_z = "recover",
recover_strand_ambig_wgt = T
# lasso/elastic-net method
res <- impute_expr_z(z_snp, weight = weight, ld_R_dir = ld_R_dir,
method = NULL, outputdir = outputdir, outname = outname.e,
harmonize_z = T, harmonize_wgt = T, scale_by_ld_variance=F,
strand_ambig_action_z = "none",
recover_strand_ambig_wgt = FGWAS: UK Biobank GWAS summary statistics - European individuals
Weights: FUSION weights using top1, lasso, or elastic-net models were converted into PredictDB format and were not needed to do scaling when running ctwas.
Check convergence of parameters
cTWAS analysis on m6A alone
[1] "Check convergence for the top1 model:"
[1] "Table of group size:"
SNP gene
8713250 888
SNP gene
estimated_group_prior 2.367e-04 0.023546
estimated_group_prior_var 2.129e+01 20.092186
estimated_group_pve 1.256e-01 0.001201
attributable_group_pve 9.905e-01 0.009474
[1] "Check convergence for the lasso model:"
[1] "Table of group size:"
SNP gene
8713250 912
SNP gene
estimated_group_prior 2.295e-04 0.030160
estimated_group_prior_var 2.061e+01 25.750541
estimated_group_pve 1.178e-01 0.002024
attributable_group_pve 9.831e-01 0.016891$top1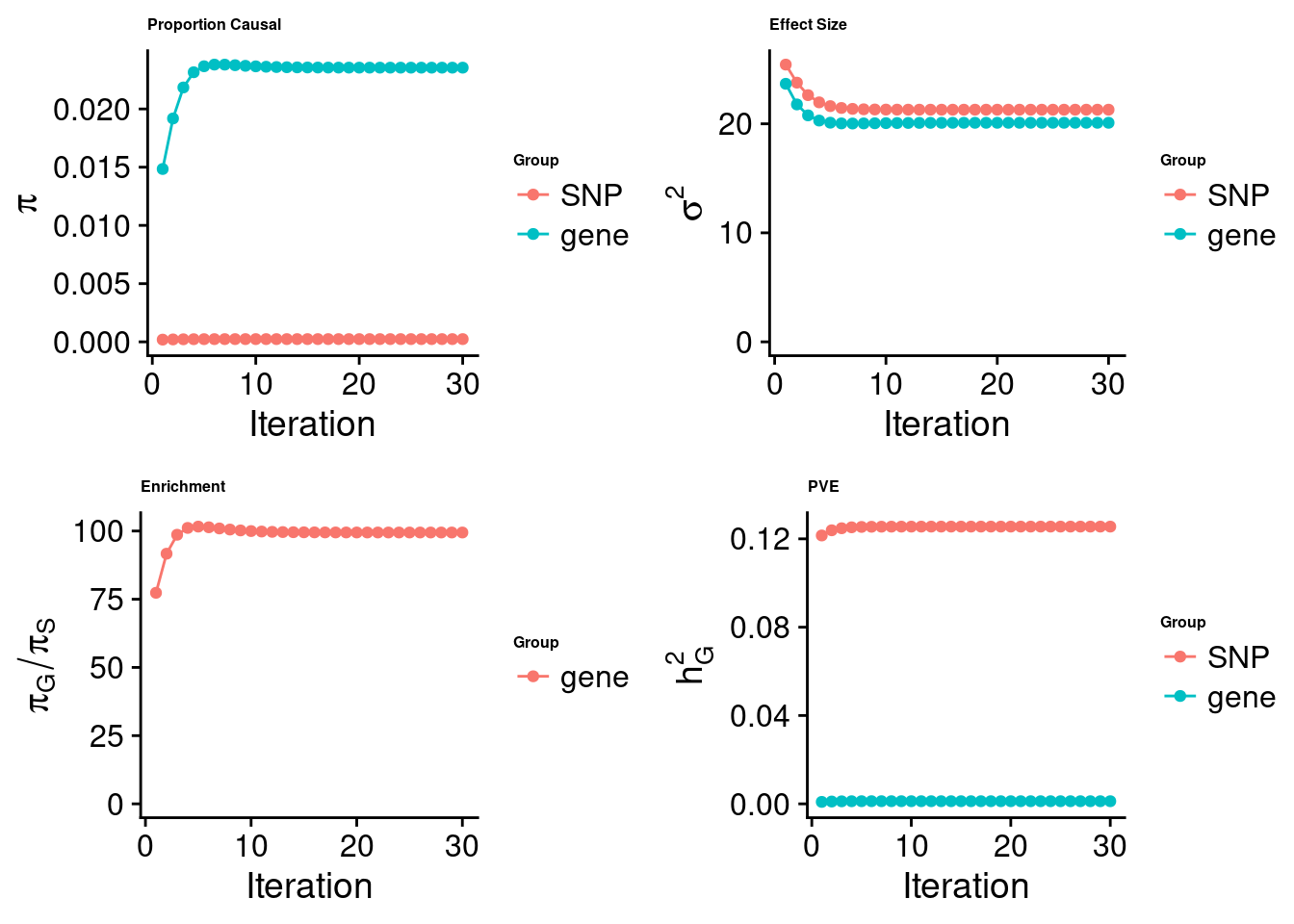
$lasso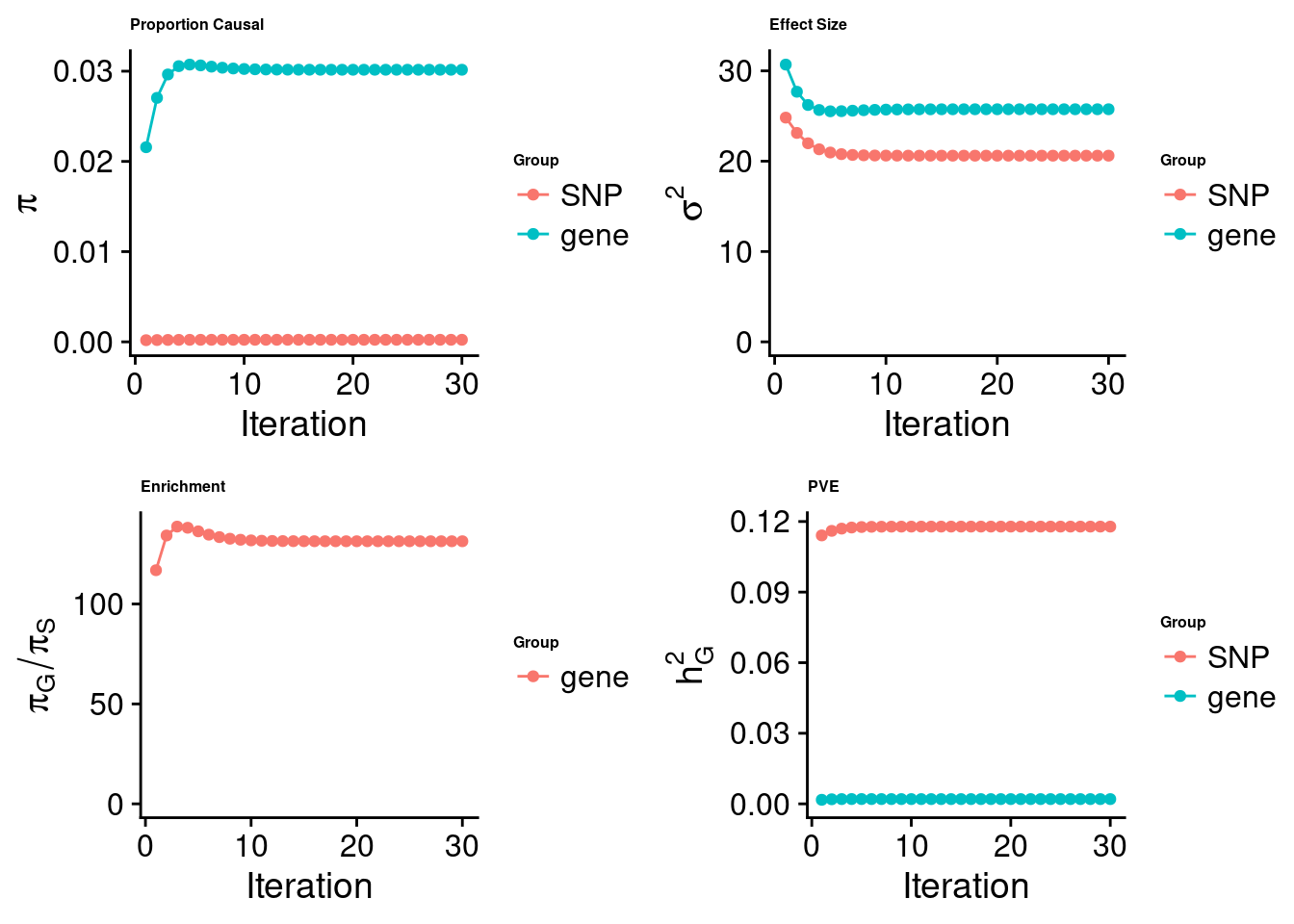
Joint analysis of expression, splicing and m6A
[1] "Check convergence for the top1 model when jointly analyzing expression, splicing and m6A:"
[1] "Table of group size before/after matching with UKBB SNPs:"
SNP eQTL sQTL m6AQTL
prior_group_size 9.324e+06 2005.0000 2191.000 918.0000
group_size 8.713e+06 1928.0000 2123.000 888.0000
percent_of_overlaps 9.345e-01 0.9616 0.969 0.9673
SNP eQTL sQTL m6AQTL
estimated_group_prior 2.259e-04 0.019133 0.013771 0.023062
estimated_group_prior_var 2.033e+01 21.099884 61.117674 17.148019
estimated_group_pve 1.144e-01 0.002225 0.005107 0.001004
attributable_group_pve 9.321e-01 0.018124 0.041608 0.008177
[1] "Check convergence for the lasso model when jointly analyzing expression, splicing and m6A:"
[1] "Table of group size before/after matching with UKBB SNPs:"
SNP eQTL sQTL m6AQTL
prior_group_size 9.324e+06 2005.0000 2191.000 918.0000
group_size 8.713e+06 1998.0000 2180.000 912.0000
percent_of_overlaps 9.345e-01 0.9965 0.995 0.9935
SNP eQTL sQTL m6AQTL
estimated_group_prior 2.012e-04 0.011316 0.010990 0.033067
estimated_group_prior_var 2.053e+01 20.406853 26.836349 23.920423
estimated_group_pve 1.029e-01 0.001319 0.001838 0.002062
attributable_group_pve 9.517e-01 0.012198 0.016998 0.019072$top1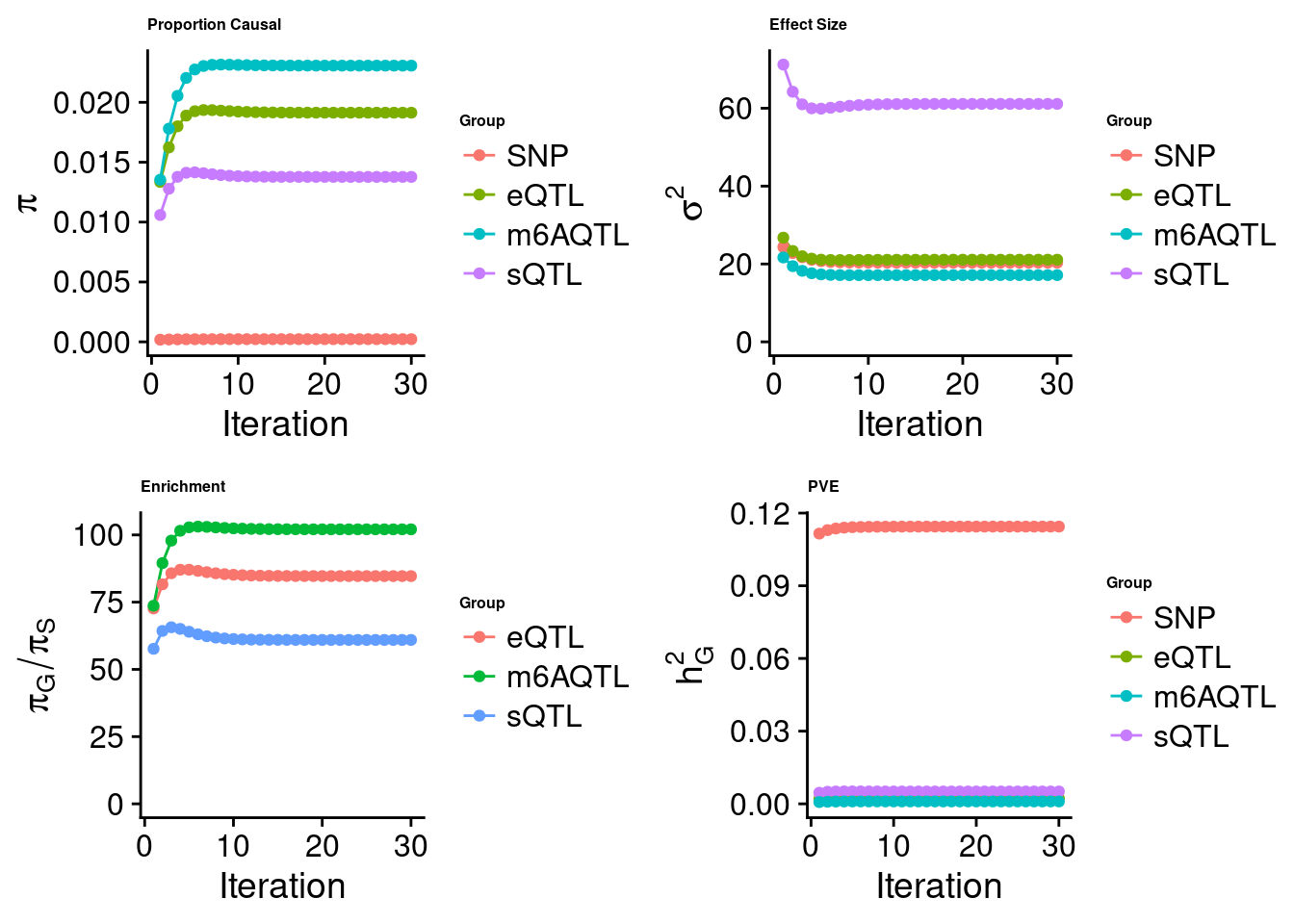
$lasso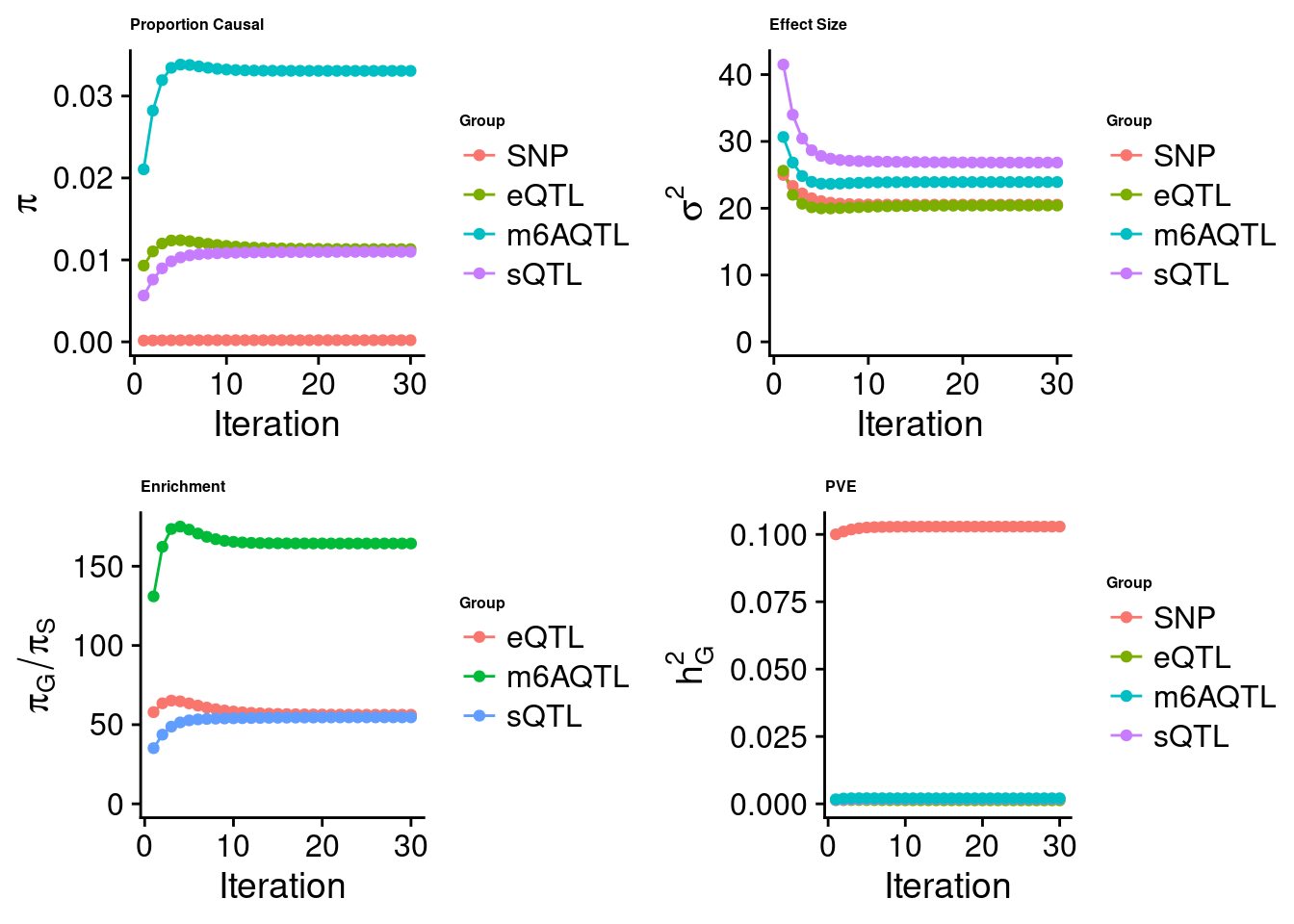
cTWAS results for individual analysis with m6A
top1 model
genename region_tag susie_pip z
1 TAP2 6_27 0.9968 -17.170
2 RANGAP1 22_17 0.9947 8.459
3 ZKSCAN5 7_61 0.9854 5.773
4 HNRNPK 9_41 0.9243 8.162
5 ADCY7 16_27 0.9120 4.410
6 MDM2 12_42 0.9089 4.085
7 ZSCAN25 7_61 0.8941 6.713
8 EPC1 10_24 0.8858 4.362
9 LAMTOR4 7_61 0.8442 4.218
10 BAZ1B 7_47 0.8115 4.511
11 THEMIS2 1_19 0.8025 7.209
12 DDX55 12_75 0.7969 6.612
13 RFTN1 3_12 0.7752 3.751
14 BRF1 14_55 0.7621 -3.816
15 RAI1 17_15 0.7489 -8.341
16 SLC25A11 17_5 0.7008 -5.529
17 GIT1 17_18 0.6791 -4.508
18 RNGTT 6_60 0.6765 3.507
19 TAF6L 11_35 0.6663 3.607
20 SMG9 19_30 0.6262 7.203Summing up PIPs for m6A peaks located in the same gene
Top m6A PIPs by genes
# A tibble: 21 × 2
genename total_susie_pip
<chr> <dbl>
1 TAP2 0.997
2 RANGAP1 0.995
3 ZKSCAN5 0.985
4 HNRNPK 0.924
5 ADCY7 0.912
6 EPC1 0.911
7 MDM2 0.909
8 ZSCAN25 0.894
9 LAMTOR4 0.871
10 BAZ1B 0.812
# ℹ 11 more rowscTWAS results for joint analysis using a lasso model
Top m6A modification pip
Top expression/splicing/m6A units
For m6A or splicing QTLs, they are assigned to the nearest genes (m6A needs to be confirmed with Kevin).
Top SNPs or genes with PIP > 0.6
$eQTL
genename susie_pip group region_tag
1888 KDELR2 0.9925 eQTL 7_9
1988 SBF1 0.8858 eQTL 22_24
1297 APH1B 0.7581 eQTL 15_29
854 MEGF9 0.7101 eQTL 9_63
701 ENSG00000182165 0.7046 eQTL 7_55
441 ENSG00000251022 0.6958 eQTL 4_56
656 GNA12 0.6501 eQTL 7_5
1878 ENSG00000153363 0.6303 eQTL 1_110
1813 ENSG00000272578 0.6242 eQTL 22_7
$m6AQTL
genename susie_pip group region_tag
5037 TAP2 0.9969 m6AQTL 6_27
5086 RANGAP1 0.9956 m6AQTL 22_17
5051 ZKSCAN5 0.9880 m6AQTL 7_61
5077 ADCY7 0.9298 m6AQTL 16_27
5067 MDM2 0.9254 m6AQTL 12_42
5052 ZSCAN25 0.9111 m6AQTL 7_61
5058 EPC1 0.9077 m6AQTL 10_24
5027 THEMIS2 0.8327 m6AQTL 1_19
5047 BAZ1B 0.8304 m6AQTL 7_47
5075 BRF1 0.7993 m6AQTL 14_55
5055 LAMTOR4 0.7843 m6AQTL 7_61
4821 SLC25A11 0.7750 m6AQTL 17_5
4923 GMIP 0.7634 m6AQTL 19_16
4443 RNGTT 0.7098 m6AQTL 6_60
5073 DDX55 0.6788 m6AQTL 12_75
4917 ADGRE2 0.6509 m6AQTL 19_12
4651 SENP1 0.6424 m6AQTL 12_31
$sQTL
genename susie_pip group region_tag
4025 APH1A 0.9979 sQTL 1_75
4062 MYO1G 0.9646 sQTL 7_33
3946 DPM1 0.7669 sQTL 20_31
4165 NAA20 0.7480 sQTL 20_14
4042 CASP8 0.7046 sQTL 2_119
2296 RNF181 0.6692 sQTL 2_54
2360 EIF4E2 0.6470 sQTL 2_137
4091 ZDHHC6 0.6398 sQTL 10_70
3192 RELT 0.6082 sQTL 11_41Top m6A modification pip
genename region_tag susie_pip z
1 TAP2 6_27 0.9969 -17.170
2 RANGAP1 22_17 0.9956 8.459
3 ZKSCAN5 7_61 0.9880 5.773
4 ADCY7 16_27 0.9298 4.410
5 MDM2 12_42 0.9254 4.085
6 ZSCAN25 7_61 0.9111 6.713
7 EPC1 10_24 0.9077 4.362
8 THEMIS2 1_19 0.8327 7.209
9 BAZ1B 7_47 0.8304 4.511
10 BRF1 14_55 0.7993 -3.816Summing up PIPs for m6A peaks located in the same gene
Top 10 m6A PIPs by genes
# A tibble: 819 × 2
genename total_susie_pip
<chr> <dbl>
1 TAP2 0.997
2 RANGAP1 0.996
3 ZKSCAN5 0.988
4 EPC1 0.939
5 ADCY7 0.930
6 MDM2 0.925
7 ZSCAN25 0.911
8 THEMIS2 0.833
9 BAZ1B 0.830
10 LAMTOR4 0.814
# ℹ 809 more rowsTop splicing PIPs
peak_id genename pos region_tag susie_pip z
1 chr1:150240527-150241098 APH1A 150210120 1_75 0.9979 6.100
2 chr7:45009474-45009639 MYO1G 44925489 7_33 0.9646 -13.705
3 chr20:49557470-49557642 DPM1 49538693 20_31 0.7669 3.989
4 chr20:20007563-20013746 NAA20 19923000 20_14 0.7480 -4.543
5 chr2:202141827-202149539 CASP8 202143928 2_119 0.7046 9.369
6 chr2:85823772-85824227 RNF181 85818886 2_54 0.6692 5.200
7 chr2:233415454-233415880 EIF4E2 233417552 2_137 0.6470 4.427
8 chr10:114186117-114186976 ZDHHC6 114169664 10_70 0.6398 -3.927
9 chr11:73101966-73102189 RELT 73002434 11_41 0.6082 3.727
10 chr1:160250036-160250976 PEX19 160150130 1_81 0.5964 4.143Summing up PIPs for spliced introns located in the same gene
Top 10 splicing PIPs by genes
# A tibble: 10 × 2
genename total_susie_pip
<chr> <dbl>
1 APH1A 0.998
2 MYO1G 0.965
3 CNN2 0.944
4 ALDH3A2 0.874
5 DPM1 0.806
6 HNRNPK 0.784
7 NAA20 0.748
8 CASP8 0.705
9 CD46 0.697
10 RNF181 0.684Top genes by combined PIP
genename combined_pip expression_pip splicing_pip m6A_pip
1518 HNRNPK 1.021 0.000e+00 0.7844982 0.23666
186 APH1A 0.998 0.000e+00 0.9979107 0.00000
2910 TAP2 0.997 0.000e+00 0.0000000 0.99695
2435 RANGAP1 0.996 0.000e+00 0.0000000 0.99559
1626 KDELR2 0.992 9.925e-01 0.0000000 0.00000
3317 ZKSCAN5 0.988 0.000e+00 0.0000000 0.98800
1957 MYO1G 0.965 0.000e+00 0.9645679 0.00000
567 CNN2 0.944 0.000e+00 0.9440766 0.00000
1239 EPC1 0.939 0.000e+00 0.0000000 0.93925
86 ADCY7 0.930 0.000e+00 0.0000000 0.92984
1792 MDM2 0.929 0.000e+00 0.0038008 0.92544
3397 ZSCAN25 0.911 0.000e+00 0.0000000 0.91106
2620 SBF1 0.886 8.858e-01 0.0000000 0.00000
133 ALDH3A2 0.874 0.000e+00 0.8736031 0.00000
1670 LAMTOR4 0.868 6.078e-03 0.0485338 0.81372
2962 THEMIS2 0.833 0.000e+00 0.0000000 0.83269
284 BAZ1B 0.830 0.000e+00 0.0000000 0.83038
1394 GMIP 0.815 0.000e+00 0.0516449 0.76338
756 DPM1 0.806 0.000e+00 0.8056706 0.00000
324 BRF1 0.799 0.000e+00 0.0000000 0.79935
2703 SLC25A11 0.775 0.000e+00 0.0000000 0.77499
187 APH1B 0.758 7.581e-01 0.0000000 0.00000
1962 NAA20 0.748 0.000e+00 0.7480051 0.00000
2758 SMG9 0.722 1.236e-01 0.0000000 0.59878
1396 GNA12 0.714 6.501e-01 0.0000000 0.06393
1810 MEGF9 0.710 7.101e-01 0.0000000 0.00000
2528 RNGTT 0.710 0.000e+00 0.0000000 0.70979
413 CASP8 0.705 0.000e+00 0.7046435 0.00000
876 ENSG00000182165 0.705 7.046e-01 0.0000000 0.00000
471 CD46 0.697 0.000e+00 0.6969764 0.00000
1887 MRPL14 0.697 1.192e-01 0.0000000 0.57744
1062 ENSG00000251022 0.696 6.958e-01 0.0000000 0.00000
2517 RNF181 0.684 0.000e+00 0.6838361 0.00000
694 DDX55 0.679 3.132e-05 0.0002274 0.67884
2481 RFTN1 0.660 0.000e+00 0.0922329 0.56765
90 ADGRE2 0.651 0.000e+00 0.0000000 0.65092
822 EIF4E2 0.647 0.000e+00 0.6470154 0.00000
2648 SENP1 0.642 5.940e-05 0.0000000 0.64241
3302 ZDHHC6 0.640 0.000e+00 0.6398440 0.00000
860 ENSG00000153363 0.630 6.303e-01 0.0000000 0.00000
region_tag
1518 9_41
186 1_75
2910 6_27
2435 22_17
1626 7_9
3317 7_61
1957 7_33
567 19_2
1239 10_24
86 16_27
1792 12_42
3397 7_61
2620 22_24
133 17_16
1670 7_61
2962 1_19
284 7_47
1394 19_16
756 20_31
324 14_55
2703 17_5
187 15_29
1962 20_14
2758 19_30
1396 7_5
1810 9_63
2528 6_60
413 2_119
876 7_55
471 1_107
1887 6_34
1062 4_56
2517 2_54
694 12_75
2481 3_12
90 19_12
822 2_137
2648 12_31
3302 10_70
860 1_110Loading required package: gridWarning: replacing previous import 'utils::download.file' by
'restfulr::download.file' when loading 'rtracklayer'Locus plots for specific examples
genename combined_pip expression_pip splicing_pip m6A_pip region_tag
1509 HMGCR 0.06 0 0 0.06028 5_44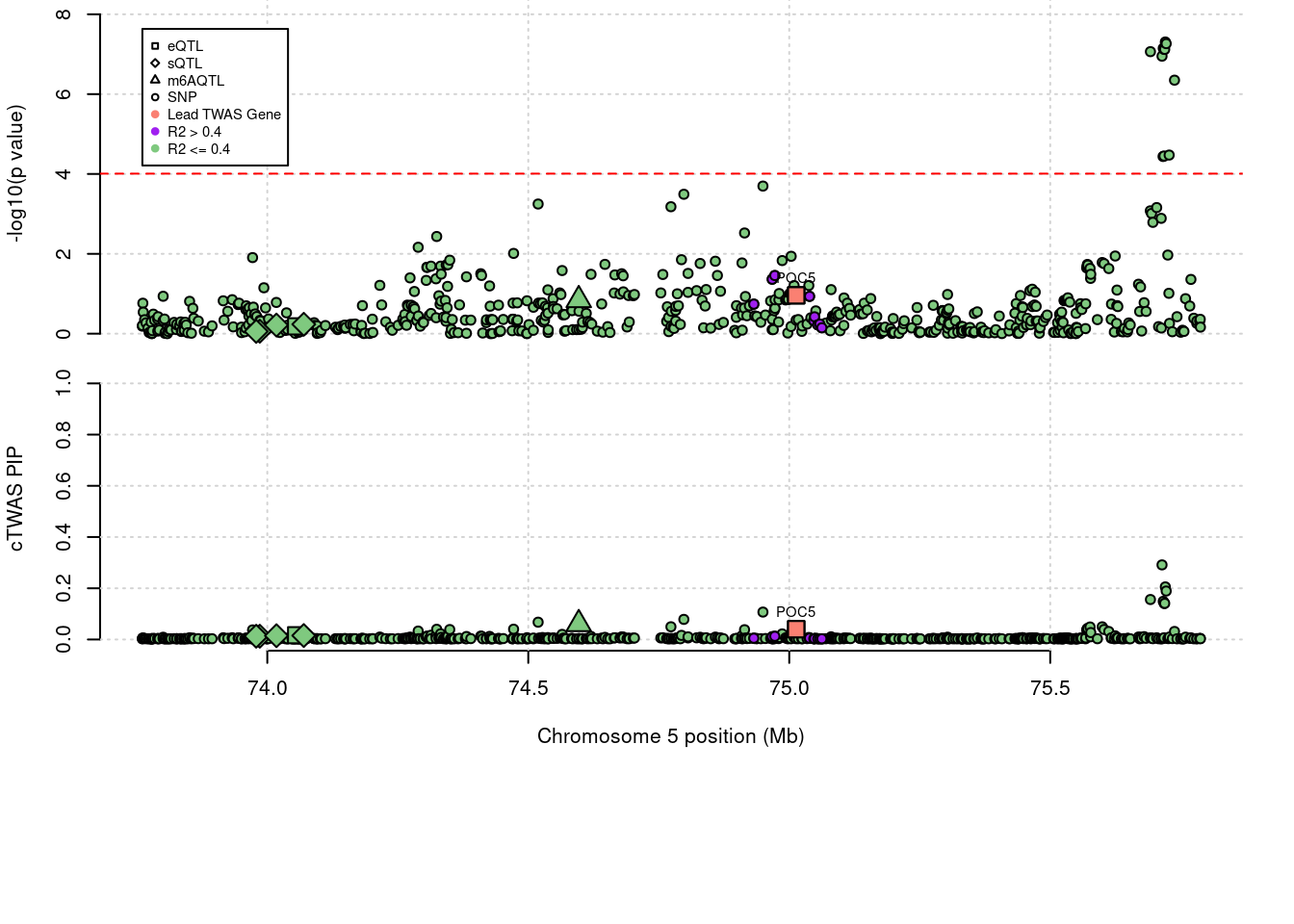
R version 4.2.0 (2022-04-22)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=C LC_MONETARY=C LC_MESSAGES=C
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] biomaRt_2.52.0 Gviz_1.40.1 cowplot_1.1.1
[4] ggplot2_3.4.3 GenomicRanges_1.48.0 GenomeInfoDb_1.32.2
[7] IRanges_2.30.1 S4Vectors_0.34.0 BiocGenerics_0.42.0
[10] ctwas_0.1.38 dplyr_1.1.2 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] colorspace_2.1-0 deldir_1.0-6
[3] rjson_0.2.21 rprojroot_2.0.3
[5] biovizBase_1.44.0 htmlTable_2.4.0
[7] XVector_0.36.0 base64enc_0.1-3
[9] fs_1.6.3 dichromat_2.0-0.1
[11] rstudioapi_0.15.0 farver_2.1.1
[13] bit64_4.0.5 AnnotationDbi_1.58.0
[15] fansi_1.0.4 xml2_1.3.3
[17] codetools_0.2-18 logging_0.10-108
[19] cachem_1.0.8 knitr_1.39
[21] Formula_1.2-4 jsonlite_1.8.7
[23] Rsamtools_2.12.0 cluster_2.1.3
[25] dbplyr_2.3.3 png_0.1-7
[27] compiler_4.2.0 httr_1.4.6
[29] backports_1.4.1 lazyeval_0.2.2
[31] Matrix_1.6-1 fastmap_1.1.1
[33] cli_3.6.1 later_1.3.0
[35] htmltools_0.5.2 prettyunits_1.1.1
[37] tools_4.2.0 gtable_0.3.3
[39] glue_1.6.2 GenomeInfoDbData_1.2.8
[41] rappdirs_0.3.3 Rcpp_1.0.11
[43] Biobase_2.56.0 jquerylib_0.1.4
[45] vctrs_0.6.3 Biostrings_2.64.0
[47] rtracklayer_1.56.0 iterators_1.0.14
[49] xfun_0.30 stringr_1.5.0
[51] ps_1.7.0 lifecycle_1.0.3
[53] ensembldb_2.20.2 restfulr_0.0.14
[55] XML_3.99-0.14 getPass_0.2-2
[57] zlibbioc_1.42.0 scales_1.2.1
[59] BSgenome_1.64.0 VariantAnnotation_1.42.1
[61] ProtGenerics_1.28.0 hms_1.1.3
[63] promises_1.2.0.1 MatrixGenerics_1.8.0
[65] parallel_4.2.0 SummarizedExperiment_1.26.1
[67] AnnotationFilter_1.20.0 RColorBrewer_1.1-3
[69] yaml_2.3.5 curl_5.0.2
[71] memoise_2.0.1 gridExtra_2.3
[73] sass_0.4.1 rpart_4.1.16
[75] latticeExtra_0.6-30 stringi_1.7.12
[77] RSQLite_2.3.1 highr_0.9
[79] BiocIO_1.6.0 foreach_1.5.2
[81] checkmate_2.1.0 GenomicFeatures_1.48.4
[83] filelock_1.0.2 BiocParallel_1.30.3
[85] rlang_1.1.1 pkgconfig_2.0.3
[87] matrixStats_0.62.0 bitops_1.0-7
[89] evaluate_0.15 lattice_0.20-45
[91] htmlwidgets_1.5.4 GenomicAlignments_1.32.0
[93] labeling_0.4.2 bit_4.0.5
[95] processx_3.8.0 tidyselect_1.2.0
[97] magrittr_2.0.3 R6_2.5.1
[99] generics_0.1.3 Hmisc_5.1-0
[101] DelayedArray_0.22.0 DBI_1.1.3
[103] pgenlibr_0.3.6 pillar_1.9.0
[105] whisker_0.4 foreign_0.8-82
[107] withr_2.5.0 KEGGREST_1.36.2
[109] RCurl_1.98-1.7 nnet_7.3-17
[111] tibble_3.2.1 crayon_1.5.2
[113] interp_1.1-4 utf8_1.2.3
[115] BiocFileCache_2.4.0 rmarkdown_2.14
[117] jpeg_0.1-10 progress_1.2.2
[119] data.table_1.14.8 blob_1.2.4
[121] callr_3.7.3 git2r_0.30.1
[123] digest_0.6.33 httpuv_1.6.5
[125] munsell_0.5.0 bslib_0.3.1